As the number of experimentally solved RNA-containing structures grows, it is becoming increasingly important to characterize the geometric features of the molecules consistently and efficiently. Existing RNA bioinformatics tools are fragmented, and suffer in either scope or usability.
DSSR, an integrated software tool for
Dissecting the
Spatial
Structure of
RNA, has been designed from ground up to
streamline the analyses of three-dimensional RNA structures. This new program consolidates, refines, and significantly extends the functionality of 3DNA for RNA structural analysis.
Starting from an RNA structure in PDB or PDBx/mmCIF format, DSSR employs a set of simple geometric criteria to identify all existent base pairs (bp): either canonical Watson-Crick and wobble pairs or non-canonical pairs with at least one hydrogen bond. The latter pairs may include normal or modified bases, regardless of tautomeric or protonation state. DSSR uses the six standard rigid-body bp parameters (shear, stretch, stagger, propeller, buckle, and opening) to rigorously quantify the spatial disposition of any two interacting bases. Where applicable, the program also denotes a bp by common names, the Saenger classification scheme of 28 H-bonding types, and the Leontis-Westhof nomenclature of 12 basic geometric classes.
DSSR detects multiplets (triplets or higher-order base associations) by searching horizontally in the plane of the associated bp for further H-bonding interactions. The program determines double-helical regions by exploring vertically in the neighborhood of selected bps for base-stacking interactions, regardless of backbone connection (e.g., coaxial stacking of helices). DSSR then identifies hairpin loops, bulges, internal loops, and multi-branch loops (junctions), and recognizes the existence of pseudo-knots. The program outputs RNA secondary structure in dot-bracket notation (dbn), connectivity table (.ct) and CRW bpseq formats that can be fed directly into visualization tools (such as VARNA).
DSSR classifies dinucleotide steps into the most common A-, B-, or Z-form double helices, calculates commonly used backbone torsion angles, and assigns the consensus RNA backbone suite names. The program also identifies A-minor interactions, ribose zippers, G quartets, kissing loops, U-turns, and kink-turns. Furthermore, it reports non-pairing interactions (H-bonding or base-stacking) between two nucleotides, and contacts involving phosphate groups.
A simple
web interface and a comprehensive
user manual are available. Supported by Dr. Robert Hanson,
DSSR has recently been integrated into Jmol, a popular molecular graphics program. DSSR-related news and information can be found on the
3DNA homepage. Questions and suggestions are always welcome on the
3DNA forum.
Give DSSR a try, compare it with similar tools in terms of usability, functionality and support, and see the differences!As of version 2.0, DSSR has been
licensed by Columbia University.
List of users who has helped improve DSSR by reporting bugs, making comments/suggestions etc:
jyvdf3asdg2;
kailsen;
MarcParisien;
jctoledo;
Auffinger;
febos;
acolasanti;
hansonr;
cllawson;
Sylverlin;
cigdem;
lvelve0901;
meier74;
jms89;
chemikeris;
Bernhard10;
rcsb_pdb;
Marcel Heinz;
lijun;
tctcab;
brinda.vallat-- Xiang-Jun
Note:
please start a new topic with a more specific title; do not post directly below this announcement.
Here are some sample runs (see
x3dna-dssr -h for more info),
x3dna-dssr -i=1msy.pdb -o=1msy.out # 27 nts
x3dna-dssr --input=1msy.pdb --output=1msy.out # as as above
x3dna-dssr -i=1msy.pdb --json -o=1msy-dssr.json # parameters exported in JSON format
x3dna-dssr -i=1ehz.pdb -o=1ehz.out # tRNA, 76 nts
x3dna-dssr -i=1jj2.pdb -o=1jj2.out # rRNA, 2876 nts
Example #1: GUAA tetraloop mutant of Sarcin/Ricin domain from
E. Coli 23 S rRNA (
1msy)
Run: x3dna-dssr -i=1msy.pdb --time-stamp=off -o=1msy.out --non-pair --u-turn
****************************************************************************
DSSR: an Integrated Software Tool for
Dissecting the Spatial Structure of RNA
v1.9.10-2020apr23, by xiangjun@x3dna.org
DSSR has been made possible by the NIH grant R01GM096889 (to X.J.Lu).
It is being actively maintained and developed. As always, I greatly
appreciate your feedback. Please report all DSSR-related issues on
the 3DNA Forum (forum.x3dna.org). I strive to respond promptly to any
questions posted there. DSSR is free of charge for NON-COMMERCIAL
purposes, and it comes with ABSOLUTELY NO WARRANTY.
****************************************************************************
Note: By default, each nucleotide is identified by chainId.name#. So a
common case would be B.A1689, meaning adenosine #1689 on chain B.
One-letter base names for modified nucleotides are put in lower
case (e.g., 'c' for 5MC). For further information about the output
notation, please refer to the DSSR User Manual.
Questions and suggestions are *always* welcome on the 3DNA Forum.
Command: x3dna-dssr -i=1msy.pdb --u-turn --non-pair -o=1msy.out
File name: 1msy.pdb
no. of DNA/RNA chains: 1 [A=27]
no. of nucleotides: 27
no. of atoms: 685
no. of waters: 109
no. of metals: 0
****************************************************************************
List of 13 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 A.U2647 A.G2673 U-G -- n/a cWW cW-W
2 A.G2648 A.U2672 G-U Wobble 28-XXVIII cWW cW-W
3 A.C2649 A.G2671 C-G WC 19-XIX cWW cW-W
4 A.U2650 A.A2670 U-A WC 20-XX cWW cW-W
5 A.C2651 A.G2669 C-G WC 19-XIX cWW cW-W
6 A.C2652 A.G2668 C-G WC 19-XIX cWW cW-W
7 A.U2653 A.C2667 U-C -- n/a tW. tW-.
8 A.A2654 A.C2666 A+C -- n/a tHH tM+M
9 A.G2655 A.U2656 G+U Platform n/a cSH cm+M
10 A.U2656 A.A2665 U-A rHoogsteen 24-XXIV tWH tW-M
11 A.A2657 A.G2664 A-G Sheared 11-XI tHS tM-m
12 A.C2658 A.G2663 C-G WC 19-XIX cWW cW-W
13 A.G2659 A.A2662 G-A Sheared 11-XI tSH tm-M
****************************************************************************
List of 1 multiplet
1 nts=3 GUA A.G2655,A.U2656,A.A2665
****************************************************************************
List of 1 helix
Note: a helix is defined by base-stacking interactions, regardless of bp
type and backbone connectivity, and may contain more than one stem.
helix#number[stems-contained] bps=number-of-base-pairs in the helix
bp-type: '|' for a canonical WC/wobble pair, '.' otherwise
helix-form: classification of a dinucleotide step comprising the bp
above the given designation and the bp that follows it. Types
include 'A', 'B' or 'Z' for the common A-, B- and Z-form helices,
'.' for an unclassified step, and 'x' for a step without a
continuous backbone.
--------------------------------------------------------------------
helix#1[1] bps=12
strand-1 5'-UGCUCCUAUACG-3'
bp-type .|||||....|.
strand-2 3'-GUGAGGCCAGGA-5'
helix-form ..AAA..x...
1 A.U2647 A.G2673 U-G -- n/a cWW cW-W
2 A.G2648 A.U2672 G-U Wobble 28-XXVIII cWW cW-W
3 A.C2649 A.G2671 C-G WC 19-XIX cWW cW-W
4 A.U2650 A.A2670 U-A WC 20-XX cWW cW-W
5 A.C2651 A.G2669 C-G WC 19-XIX cWW cW-W
6 A.C2652 A.G2668 C-G WC 19-XIX cWW cW-W
7 A.U2653 A.C2667 U-C -- n/a tW. tW-.
8 A.A2654 A.C2666 A+C -- n/a tHH tM+M
9 A.U2656 A.A2665 U-A rHoogsteen 24-XXIV tWH tW-M
10 A.A2657 A.G2664 A-G Sheared 11-XI tHS tM-m
11 A.C2658 A.G2663 C-G WC 19-XIX cWW cW-W
12 A.G2659 A.A2662 G-A Sheared 11-XI tSH tm-M
****************************************************************************
List of 1 stem
Note: a stem is defined as a helix consisting of only canonical WC/wobble
pairs, with a continuous backbone.
stem#number[#helix-number containing this stem]
Other terms are defined as in the above Helix section.
--------------------------------------------------------------------
stem#1[#1] bps=5
strand-1 5'-GCUCC-3'
bp-type |||||
strand-2 3'-UGAGG-5'
helix-form .AAA
1 A.G2648 A.U2672 G-U Wobble 28-XXVIII cWW cW-W
2 A.C2649 A.G2671 C-G WC 19-XIX cWW cW-W
3 A.U2650 A.A2670 U-A WC 20-XX cWW cW-W
4 A.C2651 A.G2669 C-G WC 19-XIX cWW cW-W
5 A.C2652 A.G2668 C-G WC 19-XIX cWW cW-W
****************************************************************************
List of 1 isolated WC/wobble pair
Note: isolated WC/wobble pairs are assigned negative indices to
differentiate them from the stem numbers, which are positive.
--------------------------------------------------------------------
[#1] -1 A.C2658 A.G2663 C-G WC 19-XIX cWW cW-W
****************************************************************************
List of 30 non-pairing interactions
1 A.U2647 A.G2648 stacking: 1.0(0.5)--pm(>>,forward) interBase-angle=6 connected min_baseDist=3.26
2 A.G2648 A.C2649 stacking: 7.3(4.6)--pm(>>,forward) interBase-angle=5 connected min_baseDist=3.30
3 A.G2648 A.G2673 stacking: 2.0(0.2)--mm(<>,outward) interBase-angle=2 min_baseDist=3.28
4 A.C2649 A.U2650 stacking: 2.8(1.1)--pm(>>,forward) interBase-angle=9 connected min_baseDist=3.09
5 A.U2650 A.C2651 stacking: 0.6(0.0)--pm(>>,forward) interBase-angle=7 connected min_baseDist=3.30
6 A.C2651 A.C2652 stacking: 0.5(0.1)--pm(>>,forward) interBase-angle=12 connected min_baseDist=3.30
7 A.C2652 A.U2653 stacking: 5.2(2.6)--pm(>>,forward) interBase-angle=13 connected min_baseDist=3.43
8 A.C2652 A.G2669 stacking: 0.2(0.0)--mm(<>,outward) interBase-angle=7 min_baseDist=3.22
9 A.U2653 A.A2654 stacking: 3.3(2.0)--pp(><,inward) interBase-angle=13 H-bonds[1]: "OP2-O2'(hydroxyl)[2.62]" connected min_baseDist=3.23
10 A.A2654 A.U2656 stacking: 3.7(1.1)--mm(<>,outward) interBase-angle=1 H-bonds[1]: "O4'*O4'[3.05]" min_baseDist=3.45
11 A.G2655 A.G2664 stacking: 4.4(2.2)--pp(><,inward) interBase-angle=10 H-bonds[2]: "O2'(hydroxyl)-O6(carbonyl)[3.09],O2'(hydroxyl)-N1(imino)[3.34]" min_baseDist=3.37
12 A.G2655 A.A2665 interBase-angle=21 H-bonds[2]: "N1(imino)-OP2[2.77],N2(amino)-O5'[2.89]" min_baseDist=4.79
13 A.U2656 A.G2664 interBase-angle=7 H-bonds[2]: "OP2-N1(imino)[3.04],OP2-N2(amino)[2.94]" min_baseDist=3.36
14 A.A2657 A.C2658 stacking: 6.7(2.6)--pm(>>,forward) interBase-angle=4 connected min_baseDist=3.46
15 A.A2657 A.A2665 stacking: 3.7(3.3)--mm(<>,outward) interBase-angle=11 min_baseDist=3.29
16 A.C2658 A.G2659 stacking: 0.4(0.1)--pm(>>,forward) interBase-angle=10 connected min_baseDist=3.34
17 A.G2659 A.A2661 interBase-angle=31 H-bonds[2]: "O2'(hydroxyl)-N7[2.60],O2'(hydroxyl)-N6(amino)[3.26]" min_baseDist=3.97
18 A.G2659 A.G2663 stacking: 3.9(1.2)--mm(<>,outward) interBase-angle=4 min_baseDist=3.35
19 A.U2660 A.A2661 stacking: 7.5(4.2)--pm(>>,forward) interBase-angle=17 connected min_baseDist=3.26
20 A.A2661 A.A2662 stacking: 6.3(4.4)--pm(>>,forward) interBase-angle=19 connected min_baseDist=3.38
21 A.G2663 A.G2664 stacking: 2.7(0.6)--pm(>>,forward) interBase-angle=8 connected min_baseDist=3.38
22 A.G2664 A.A2665 interBase-angle=14 H-bonds[1]: "O2'(hydroxyl)-O4'[2.75]" connected min_baseDist=5.83
23 A.A2665 A.C2666 stacking: 1.6(1.1)--pm(>>,forward) interBase-angle=10 connected min_baseDist=3.18
24 A.C2666 A.C2667 stacking: 4.3(2.1)--pm(>>,forward) interBase-angle=8 connected min_baseDist=3.35
25 A.C2667 A.G2668 stacking: 3.1(1.0)--pm(>>,forward) interBase-angle=7 connected min_baseDist=3.38
26 A.G2668 A.G2669 stacking: 4.3(3.0)--pm(>>,forward) interBase-angle=4 connected min_baseDist=3.28
27 A.G2669 A.A2670 stacking: 4.3(2.9)--pm(>>,forward) interBase-angle=4 connected min_baseDist=3.29
28 A.A2670 A.G2671 stacking: 1.5(1.5)--pm(>>,forward) interBase-angle=6 connected min_baseDist=3.24
29 A.G2671 A.U2672 stacking: 7.4(4.0)--pm(>>,forward) interBase-angle=10 connected min_baseDist=3.22
30 A.U2672 A.G2673 interBase-angle=11 H-bonds[1]: "O2'(hydroxyl)-O4'[3.37]" connected min_baseDist=3.61
****************************************************************************
List of 5 stacks
Note: a stack is an ordered list of nucleotides assembled together via
base-stacking interactions, regardless of backbone connectivity.
Stacking interactions within a stem are *not* included.
1 nts=2 GG A.G2648,A.G2673
2 nts=3 UAA A.U2660,A.A2661,A.A2662
3 nts=4 CUAU A.C2652,A.U2653,A.A2654,A.U2656
4 nts=4 GGGG A.G2655,A.G2664,A.G2663,A.G2659
5 nts=6 CAACCG A.C2658,A.A2657,A.A2665,A.C2666,A.C2667,A.G2668
****************************************************************************
List of 2 atom-base capping interactions
dv: vertical distance of the atom above the nucleotide base
-----------------------------------------------------------
type atom nt dv
1 phosphate OP2@A.A2661 A.G2659 3.04
2 sugar O4'@A.G2664 A.G2663 3.48
****************************************************************************
Note: for the various types of loops listed below, numbers within the first
set of brackets are the number of loop nts, and numbers in the second
set of brackets are the identities of the stems (positive number) or
isolated WC/wobble pairs (negative numbers) to which they are linked.
****************************************************************************
List of 1 hairpin loop
1 hairpin loop: nts=6; [4]; linked by [#-1]
summary: [1] 4 [A.2658 A.2663] 1
nts=6 CGUAAG A.C2658,A.G2659,A.U2660,A.A2661,A.A2662,A.G2663
nts=4 GUAA A.G2659,A.U2660,A.A2661,A.A2662
****************************************************************************
List of 1 internal loop
1 asymmetric internal loop: nts=13; [5,4]; linked by [#1,#-1]
summary: [2] 5 4 [A.2652 A.2668 A.2658 A.2663] 5 1
nts=13 CUAGUACGGACCG A.C2652,A.U2653,A.A2654,A.G2655,A.U2656,A.A2657,A.C2658,A.G2663,A.G2664,A.A2665,A.C2666,A.C2667,A.G2668
nts=5 UAGUA A.U2653,A.A2654,A.G2655,A.U2656,A.A2657
nts=4 GACC A.G2664,A.A2665,A.C2666,A.C2667
****************************************************************************
List of 2 non-loop single-stranded segments
1 nts=1 U A.U2647
2 nts=1 G A.G2673
****************************************************************************
List of 1 U-turn
1 A.G2659-A.A2662 H-bonds[2]: "N2(amino)-OP2[2.97],N2(amino)-N7[2.86]" nts=6 CGUAAG A.C2658,A.G2659,A.U2660,A.A2661,A.A2662,A.G2663
****************************************************************************
List of 1 splayed-apart dinucleotide
1 A.G2659 A.U2660 angle=95 distance=13.2 ratio=0.74
----------------------------------------------------------------
Summary of 1 splayed-apart unit
1 nts=2 GU A.G2659,A.U2660
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>1msy nts=27 [whole]
UGCUCCUAGUACGUAAGGACCGGAGUG
.(((((.....(....)....))))).
>1msy-A #1 nts=27 0.30(2.47) [chain] RNA
UGCUCCUAGUACGUAAGGACCGGAGUG
.(((((.....(....)....))))).
****************************************************************************
Summary of structural features of 27 nucleotides
Note: the first five columns are: (1) serial number, (2) one-letter
shorthand name, (3) dbn, (4) id string, (5) rmsd (~zero) of base
ring atoms fitted against those in a standard base reference
frame. The sixth (last) column contains a comma-separated list of
features whose meanings are mostly self-explanatory, except for:
turn: angle C1'(i-1)--C1'(i)--C1'(i+1) < 90 degrees
break: no backbone linkage between O3'(i-1) and P(i)
1 U . A.U2647 0.011 anti,~C3'-endo,non-canonical,non-pair-contact,helix-end,ss-non-loop
2 G ( A.G2648 0.012 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end
3 C ( A.C2649 0.019 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem
4 U ( A.U2650 0.019 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem
5 C ( A.C2651 0.024 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem
6 C ( A.C2652 0.032 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,internal-loop
7 U . A.U2653 0.019 anti,~C3'-endo,non-canonical,non-pair-contact,helix,internal-loop,phosphate
8 A . A.A2654 0.019 anti,~C2'-endo,BII,non-canonical,non-pair-contact,helix,internal-loop
9 G . A.G2655 0.022 turn,anti,~C2'-endo,non-canonical,non-pair-contact,multiplet,internal-loop
10 U . A.U2656 0.020 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,multiplet,internal-loop,phosphate
11 A . A.A2657 0.023 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,internal-loop
12 C ( A.C2658 0.013 anti,~C3'-endo,BI,isolated-canonical,non-pair-contact,helix,hairpin-loop,internal-loop
13 G . A.G2659 0.033 u-turn,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix-end,hairpin-loop,cap-acceptor,splayed-apart
14 U . A.U2660 0.020 turn,u-turn,anti,~C3'-endo,non-pair-contact,hairpin-loop,splayed-apart
15 A . A.A2661 0.015 u-turn,anti,~C3'-endo,BI,non-pair-contact,hairpin-loop,cap-donor,phosphate
16 A . A.A2662 0.010 u-turn,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix-end,hairpin-loop,phosphate
17 G ) A.G2663 0.019 anti,~C3'-endo,BI,isolated-canonical,non-pair-contact,helix,hairpin-loop,internal-loop,cap-acceptor
18 G . A.G2664 0.014 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,internal-loop,cap-donor
19 A . A.A2665 0.014 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,multiplet,internal-loop,phosphate
20 C . A.C2666 0.016 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,internal-loop,phosphate
21 C . A.C2667 0.029 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,internal-loop
22 G ) A.G2668 0.012 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,internal-loop
23 G ) A.G2669 0.020 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem
24 A ) A.A2670 0.019 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem
25 G ) A.G2671 0.023 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem
26 U ) A.U2672 0.024 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end
27 G . A.G2673 0.010 anti,~C3'-endo,non-canonical,non-pair-contact,helix-end,ss-non-loop
****************************************************************************
List of 14 additional files
1 dssr-pairs.pdb -- an ensemble of base pairs
2 dssr-multiplets.pdb -- an ensemble of multiplets
3 dssr-stems.pdb -- an ensemble of stems
4 dssr-helices.pdb -- an ensemble of helices (coaxial stacking)
5 dssr-hairpins.pdb -- an ensemble of hairpin loops
6 dssr-iloops.pdb -- an ensemble of internal loops
7 dssr-2ndstrs.bpseq -- secondary structure in bpseq format
8 dssr-2ndstrs.ct -- secondary structure in connectivity table format
9 dssr-2ndstrs.dbn -- secondary structure in dot-bracket notation
10 dssr-torsions.txt -- backbone torsion angles and suite names
11 dssr-splays.pdb -- an ensemble of splayed-apart units
12 dssr-Uturns.pdb -- an ensemble of U-turn motifs
13 dssr-stacks.pdb -- an ensemble of stacks
14 dssr-atom2bases.pdb -- an ensemble of atom-base stacking interactions
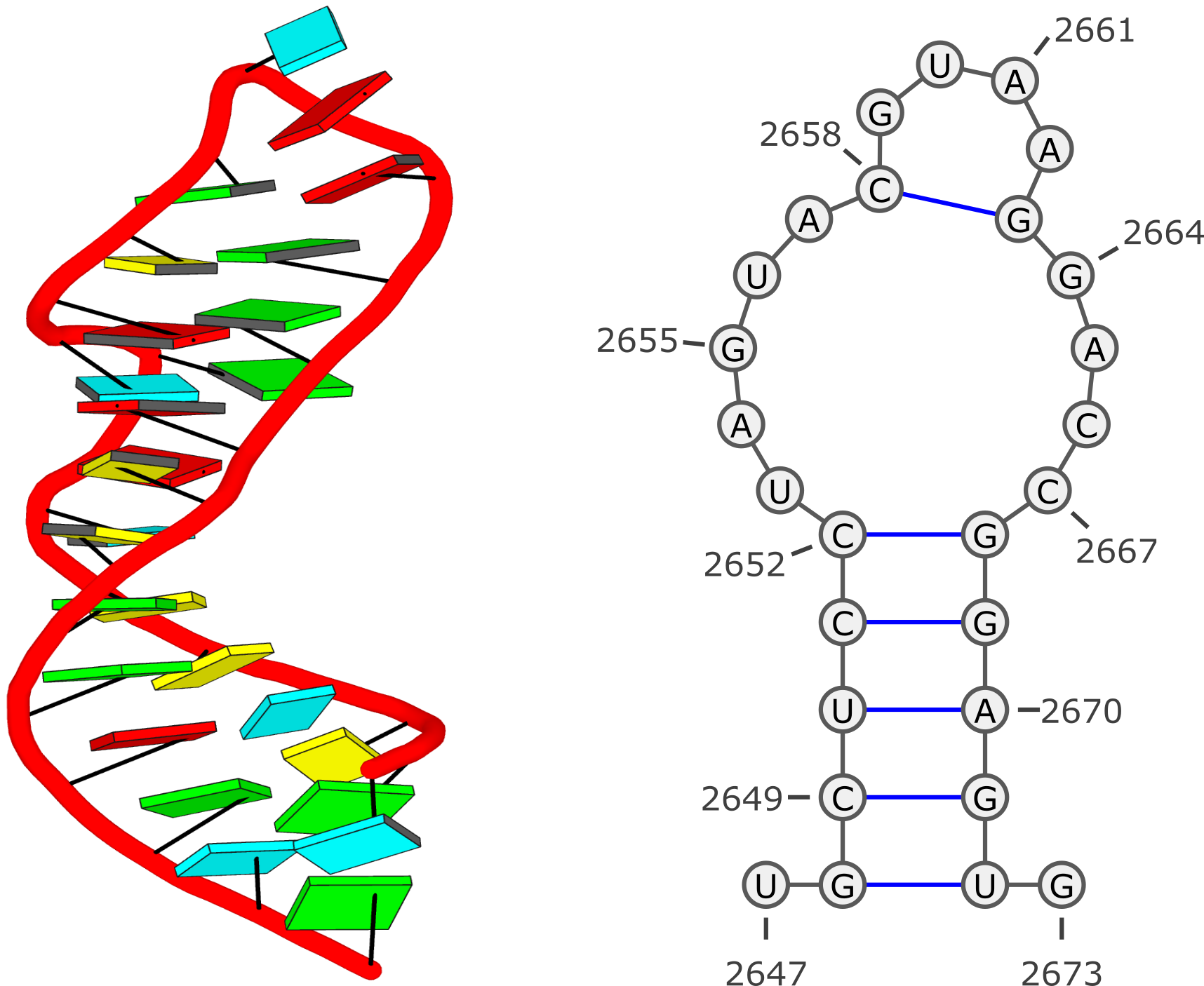
: The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution (
1ehz)
Run: x3dna-dssr -i=1ehz.pdb --time-stamp=off -o=1ehz.out --po4 --u-turn
****************************************************************************
DSSR: an Integrated Software Tool for
Dissecting the Spatial Structure of RNA
v1.9.10-2020apr23, by xiangjun@x3dna.org
DSSR has been made possible by the NIH grant R01GM096889 (to X.J.Lu).
It is being actively maintained and developed. As always, I greatly
appreciate your feedback. Please report all DSSR-related issues on
the 3DNA Forum (forum.x3dna.org). I strive to respond promptly to any
questions posted there. DSSR is free of charge for NON-COMMERCIAL
purposes, and it comes with ABSOLUTELY NO WARRANTY.
****************************************************************************
Note: By default, each nucleotide is identified by chainId.name#. So a
common case would be B.A1689, meaning adenosine #1689 on chain B.
One-letter base names for modified nucleotides are put in lower
case (e.g., 'c' for 5MC). For further information about the output
notation, please refer to the DSSR User Manual.
Questions and suggestions are *always* welcome on the 3DNA Forum.
Command: x3dna-dssr -i=1ehz.pdb --u-turn --po4 -o=1ehz.out
File name: 1ehz.pdb
no. of DNA/RNA chains: 1 [A=76]
no. of nucleotides: 76
no. of atoms: 1821
no. of waters: 160
no. of metals: 9 [Mg=6,Mn=3]
****************************************************************************
List of 11 types of 14 modified nucleotides
nt count list
1 1MA-a 1 A.1MA58
2 2MG-g 1 A.2MG10
3 5MC-c 2 A.5MC40,A.5MC49
4 5MU-t 1 A.5MU54
5 7MG-g 1 A.7MG46
6 H2U-u 2 A.H2U16,A.H2U17
7 M2G-g 1 A.M2G26
8 OMC-c 1 A.OMC32
9 OMG-g 1 A.OMG34
10 PSU-P 2 A.PSU39,A.PSU55
11 YYG-g 1 A.YYG37
****************************************************************************
List of 34 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 A.G1 A.C72 G-C WC 19-XIX cWW cW-W
2 A.C2 A.G71 C-G WC 19-XIX cWW cW-W
3 A.G3 A.C70 G-C WC 19-XIX cWW cW-W
4 A.G4 A.U69 G-U Wobble 28-XXVIII cWW cW-W
5 A.A5 A.U68 A-U WC 20-XX cWW cW-W
6 A.U6 A.A67 U-A WC 20-XX cWW cW-W
7 A.U7 A.A66 U-A WC 20-XX cWW cW-W
8 A.U8 A.A14 U-A rHoogsteen 24-XXIV tWH tW-M
9 A.U8 A.A21 U+A -- n/a tSW tm+W
10 A.A9 A.A23 A+A -- 02-II tHH tM+M
11 A.2MG10 A.C25 g-C WC 19-XIX cWW cW-W
12 A.2MG10 A.G45 g+G -- n/a cHS cM+m
13 A.C11 A.G24 C-G WC 19-XIX cWW cW-W
14 A.U12 A.A23 U-A WC 20-XX cWW cW-W
15 A.C13 A.G22 C-G WC 19-XIX cWW cW-W
16 A.G15 A.C48 G+C rWC 22-XXII tWW tW+W
17 A.H2U16 A.U59 u+U -- n/a tSW tm+W
18 A.G18 A.PSU55 G+P -- n/a tWS tW+m
19 A.G19 A.C56 G-C WC 19-XIX cWW cW-W
20 A.G22 A.7MG46 G-g -- 07-VII tHW tM-W
21 A.M2G26 A.A44 g-A Imino 08-VIII cWW cW-W
22 A.C27 A.G43 C-G WC 19-XIX cWW cW-W
23 A.C28 A.G42 C-G WC 19-XIX cWW cW-W
24 A.A29 A.U41 A-U WC 20-XX cWW cW-W
25 A.G30 A.5MC40 G-c WC 19-XIX cWW cW-W
26 A.A31 A.PSU39 A-P -- n/a cWW cW-W
27 A.OMC32 A.A38 c-A -- n/a c.W c.-W
28 A.U33 A.A36 U-A -- n/a tSH tm-M
29 A.5MC49 A.G65 c-G WC 19-XIX cWW cW-W
30 A.U50 A.A64 U-A WC 20-XX cWW cW-W
31 A.G51 A.C63 G-C WC 19-XIX cWW cW-W
32 A.U52 A.A62 U-A WC 20-XX cWW cW-W
33 A.G53 A.C61 G-C WC 19-XIX cWW cW-W
34 A.5MU54 A.1MA58 t-a rHoogsteen 24-XXIV tWH tW-M
****************************************************************************
List of 4 multiplets
1 nts=3 UAA A.U8,A.A14,A.A21
2 nts=3 AUA A.A9,A.U12,A.A23
3 nts=3 gCG A.2MG10,A.C25,A.G45
4 nts=3 CGg A.C13,A.G22,A.7MG46
****************************************************************************
List of 2 helices
Note: a helix is defined by base-stacking interactions, regardless of bp
type and backbone connectivity, and may contain more than one stem.
helix#number[stems-contained] bps=number-of-base-pairs in the helix
bp-type: '|' for a canonical WC/wobble pair, '.' otherwise
helix-form: classification of a dinucleotide step comprising the bp
above the given designation and the bp that follows it. Types
include 'A', 'B' or 'Z' for the common A-, B- and Z-form helices,
'.' for an unclassified step, and 'x' for a step without a
continuous backbone.
--------------------------------------------------------------------
helix#1[2] bps=15
strand-1 5'-GCGGAUUcUGUGtPC-3'
bp-type ||||||||||||..|
strand-2 3'-CGCUUAAGACACaGG-5'
helix-form AA....xAAAAxx.
1 A.G1 A.C72 G-C WC 19-XIX cWW cW-W
2 A.C2 A.G71 C-G WC 19-XIX cWW cW-W
3 A.G3 A.C70 G-C WC 19-XIX cWW cW-W
4 A.G4 A.U69 G-U Wobble 28-XXVIII cWW cW-W
5 A.A5 A.U68 A-U WC 20-XX cWW cW-W
6 A.U6 A.A67 U-A WC 20-XX cWW cW-W
7 A.U7 A.A66 U-A WC 20-XX cWW cW-W
8 A.5MC49 A.G65 c-G WC 19-XIX cWW cW-W
9 A.U50 A.A64 U-A WC 20-XX cWW cW-W
10 A.G51 A.C63 G-C WC 19-XIX cWW cW-W
11 A.U52 A.A62 U-A WC 20-XX cWW cW-W
12 A.G53 A.C61 G-C WC 19-XIX cWW cW-W
13 A.5MU54 A.1MA58 t-a rHoogsteen 24-XXIV tWH tW-M
14 A.PSU55 A.G18 P+G -- n/a tSW tm+W
15 A.C56 A.G19 C-G WC 19-XIX cWW cW-W
--------------------------------------------------------------------------
helix#2[2] bps=15
strand-1 5'-AAPcUGGAgCUCAGu-3'
bp-type ...||||.||||...
strand-2 3'-UcAGACCgCGAGUCU-5'
helix-form x..AAAAxAA.xxx
1 A.A36 A.U33 A-U -- n/a tHS tM-m
2 A.A38 A.OMC32 A-c -- n/a cW. cW-.
3 A.PSU39 A.A31 P-A -- n/a cWW cW-W
4 A.5MC40 A.G30 c-G WC 19-XIX cWW cW-W
5 A.U41 A.A29 U-A WC 20-XX cWW cW-W
6 A.G42 A.C28 G-C WC 19-XIX cWW cW-W
7 A.G43 A.C27 G-C WC 19-XIX cWW cW-W
8 A.A44 A.M2G26 A-g Imino 08-VIII cWW cW-W
9 A.2MG10 A.C25 g-C WC 19-XIX cWW cW-W
10 A.C11 A.G24 C-G WC 19-XIX cWW cW-W
11 A.U12 A.A23 U-A WC 20-XX cWW cW-W
12 A.C13 A.G22 C-G WC 19-XIX cWW cW-W
13 A.A14 A.U8 A-U rHoogsteen 24-XXIV tHW tM-W
14 A.G15 A.C48 G+C rWC 22-XXII tWW tW+W
15 A.H2U16 A.U59 u+U -- n/a tSW tm+W
****************************************************************************
List of 4 stems
Note: a stem is defined as a helix consisting of only canonical WC/wobble
pairs, with a continuous backbone.
stem#number[#helix-number containing this stem]
Other terms are defined as in the above Helix section.
--------------------------------------------------------------------
stem#1[#1] bps=7
strand-1 5'-GCGGAUU-3'
bp-type |||||||
strand-2 3'-CGCUUAA-5'
helix-form AA....
1 A.G1 A.C72 G-C WC 19-XIX cWW cW-W
2 A.C2 A.G71 C-G WC 19-XIX cWW cW-W
3 A.G3 A.C70 G-C WC 19-XIX cWW cW-W
4 A.G4 A.U69 G-U Wobble 28-XXVIII cWW cW-W
5 A.A5 A.U68 A-U WC 20-XX cWW cW-W
6 A.U6 A.A67 U-A WC 20-XX cWW cW-W
7 A.U7 A.A66 U-A WC 20-XX cWW cW-W
--------------------------------------------------------------------------
stem#2[#2] bps=4
strand-1 5'-gCUC-3'
bp-type ||||
strand-2 3'-CGAG-5'
helix-form AA.
1 A.2MG10 A.C25 g-C WC 19-XIX cWW cW-W
2 A.C11 A.G24 C-G WC 19-XIX cWW cW-W
3 A.U12 A.A23 U-A WC 20-XX cWW cW-W
4 A.C13 A.G22 C-G WC 19-XIX cWW cW-W
--------------------------------------------------------------------------
stem#3[#2] bps=4
strand-1 5'-CCAG-3'
bp-type ||||
strand-2 3'-GGUc-5'
helix-form AAA
1 A.C27 A.G43 C-G WC 19-XIX cWW cW-W
2 A.C28 A.G42 C-G WC 19-XIX cWW cW-W
3 A.A29 A.U41 A-U WC 20-XX cWW cW-W
4 A.G30 A.5MC40 G-c WC 19-XIX cWW cW-W
--------------------------------------------------------------------------
stem#4[#1] bps=5
strand-1 5'-cUGUG-3'
bp-type |||||
strand-2 3'-GACAC-5'
helix-form AAAA
1 A.5MC49 A.G65 c-G WC 19-XIX cWW cW-W
2 A.U50 A.A64 U-A WC 20-XX cWW cW-W
3 A.G51 A.C63 G-C WC 19-XIX cWW cW-W
4 A.U52 A.A62 U-A WC 20-XX cWW cW-W
5 A.G53 A.C61 G-C WC 19-XIX cWW cW-W
****************************************************************************
List of 1 isolated WC/wobble pair
Note: isolated WC/wobble pairs are assigned negative indices to
differentiate them from the stem numbers, which are positive.
--------------------------------------------------------------------
[#1] -1 A.G19 A.C56 G-C WC 19-XIX cWW cW-W
****************************************************************************
List of 2 coaxial stacks
1 Helix#1 contains 2 stems: [#1,#4]
2 Helix#2 contains 2 stems: [#3,#2]
****************************************************************************
List of 11 stacks
Note: a stack is an ordered list of nucleotides assembled together via
base-stacking interactions, regardless of backbone connectivity.
Stacking interactions within a stem are *not* included.
1 nts=2 Uc A.U7,A.5MC49
2 nts=2 UC A.U8,A.C13
3 nts=2 GA A.G65,A.A66
4 nts=3 CgC A.C25,A.M2G26,A.C27
5 nts=3 gAC A.7MG46,A.A21,A.C48
6 nts=3 GtP A.G53,A.5MU54,A.PSU55
7 nts=4 GACC A.G1,A.A73,A.C74,A.C75
8 nts=4 GAcU A.G30,A.A31,A.OMC32,A.U33
9 nts=5 GGGaC A.G19,A.G57,A.G18,A.1MA58,A.C61
10 nts=7 gAAgAPc A.OMG34,A.A35,A.A36,A.YYG37,A.A38,A.PSU39,A.5MC40
11 nts=9 GAGAGAGUC A.G43,A.A44,A.G45,A.A9,A.G22,A.A14,A.G15,A.U59,A.C60
****************************************************************************
Nucleotides not involved in stacking interactions
nts=4 uGUA A.H2U17,A.G20,A.U47,A.A76
****************************************************************************
List of 4 atom-base capping interactions
dv: vertical distance of the atom above the nucleotide base
-----------------------------------------------------------
type atom nt dv
1 phosphate OP2@A.H2U16 A.H2U16 2.59
2 phosphate OP2@A.A35 A.U33 2.85
3 sugar O4'@A.U59 A.C48 3.10
4 phosphate OP2@A.G57 A.PSU55 2.90
****************************************************************************
Note: for the various types of loops listed below, numbers within the first
set of brackets are the number of loop nts, and numbers in the second
set of brackets are the identities of the stems (positive number) or
isolated WC/wobble pairs (negative numbers) to which they are linked.
****************************************************************************
List of 3 hairpin loops
1 hairpin loop: nts=10; [8]; linked by [#2]
summary: [1] 8 [A.13 A.22] 4
nts=10 CAGuuGGGAG A.C13,A.A14,A.G15,A.H2U16,A.H2U17,A.G18,A.G19,A.G20,A.A21,A.G22
nts=8 AGuuGGGA A.A14,A.G15,A.H2U16,A.H2U17,A.G18,A.G19,A.G20,A.A21
2 hairpin loop: nts=11; [9]; linked by [#3]
summary: [1] 9 [A.30 A.40] 4
nts=11 GAcUgAAgAPc A.G30,A.A31,A.OMC32,A.U33,A.OMG34,A.A35,A.A36,A.YYG37,A.A38,A.PSU39,A.5MC40
nts=9 AcUgAAgAP A.A31,A.OMC32,A.U33,A.OMG34,A.A35,A.A36,A.YYG37,A.A38,A.PSU39
3 hairpin loop: nts=9; [7]; linked by [#4]
summary: [1] 7 [A.53 A.61] 5
nts=9 GtPCGaUCC A.G53,A.5MU54,A.PSU55,A.C56,A.G57,A.1MA58,A.U59,A.C60,A.C61
nts=7 tPCGaUC A.5MU54,A.PSU55,A.C56,A.G57,A.1MA58,A.U59,A.C60
****************************************************************************
List of 1 junction
1 4-way junction: nts=16; [2,1,5,0]; linked by [#1,#2,#3,#4]
summary: [4] 2 1 5 0 [A.7 A.66 A.10 A.25 A.27 A.43 A.49 A.65] 7 4 4 5
nts=16 UUAgCgCGAGgUCcGA A.U7,A.U8,A.A9,A.2MG10,A.C25,A.M2G26,A.C27,A.G43,A.A44,A.G45,A.7MG46,A.U47,A.C48,A.5MC49,A.G65,A.A66
nts=2 UA A.U8,A.A9
nts=1 g A.M2G26
nts=5 AGgUC A.A44,A.G45,A.7MG46,A.U47,A.C48
nts=0
****************************************************************************
List of 1 non-loop single-stranded segment
1 nts=4 ACCA A.A73,A.C74,A.C75,A.A76
****************************************************************************
List of 1 kissing loop interaction
1 isolated-pair #-1 between hairpin loops #1 and #3
****************************************************************************
List of 2 U-turns
1 A.U33-A.A36 H-bonds[1]: "N3(imino)-OP2[2.80]" nts=6 cUgAAg A.OMC32,A.U33,A.OMG34,A.A35,A.A36,A.YYG37
2 A.PSU55-A.1MA58 H-bonds[1]: "N3(imino)-OP2[2.77]" nts=6 tPCGaU A.5MU54,A.PSU55,A.C56,A.G57,A.1MA58,A.U59
****************************************************************************
List of 18 phosphate interactions
1 A.U7 OP1-hbonds[1]: "MG@A.MG580[2.60]"
2 A.A9 OP2-hbonds[1]: "N4@A.C13[3.01]"
3 A.A14 OP2-hbonds[1]: "MG@A.MG580[1.93]"
4 A.H2U16 OP2-cap: "A.H2U16"
5 A.G18 OP1-hbonds[1]: "O2'@A.H2U17[2.97]"
6 A.G19 OP1-hbonds[2]: "N4@A.C60[3.27],MN@A.MN530[2.19]"
7 A.G20 OP1-hbonds[1]: "MG@A.MG540[2.07]"
8 A.A21 OP2-hbonds[1]: "MG@A.MG540[2.11]"
9 A.A23 OP2-hbonds[1]: "N6@A.A9[3.12]"
10 A.A35 OP2-cap: "A.U33"
11 A.A36 OP2-hbonds[1]: "N3@A.U33[2.80]"
12 A.YYG37 OP2-hbonds[1]: "MG@A.MG590[2.53]"
13 A.C48 OP2-hbonds[1]: "O2'@A.7MG46[3.55]"
14 A.5MC49 OP1-hbonds[1]: "O2'@A.C48[3.13]" OP2-hbonds[1]: "O2'@A.U7[2.68]"
15 A.U50 OP1-hbonds[1]: "O2'@A.U47[2.71]"
16 A.G57 OP2-cap: "A.PSU55"
17 A.1MA58 OP2-hbonds[1]: "N3@A.PSU55[2.77]"
18 A.C60 OP1-hbonds[1]: "N4@A.C61[3.12]" OP2-hbonds[1]: "O2'@A.1MA58[2.42]"
****************************************************************************
List of 9 splayed-apart dinucleotides
1 A.U7 A.U8 angle=127 distance=17.2 ratio=0.90
2 A.H2U16 A.H2U17 angle=146 distance=19.5 ratio=0.96
3 A.H2U17 A.G18 angle=106 distance=15.8 ratio=0.80
4 A.G19 A.G20 angle=130 distance=16.8 ratio=0.91
5 A.7MG46 A.U47 angle=139 distance=19.0 ratio=0.94
6 A.U47 A.C48 angle=157 distance=19.7 ratio=0.98
7 A.C48 A.5MC49 angle=148 distance=17.5 ratio=0.96
8 A.C60 A.C61 angle=91 distance=13.8 ratio=0.71
9 A.C75 A.A76 angle=160 distance=18.7 ratio=0.98
----------------------------------------------------------------
Summary of 6 splayed-apart units
1 nts=2 UU A.U7,A.U8
2 nts=3 uuG A.H2U16,A.H2U17,A.G18
3 nts=2 GG A.G19,A.G20
4 nts=4 gUCc A.7MG46,A.U47,A.C48,A.5MC49
5 nts=2 CC A.C60,A.C61
6 nts=2 CA A.C75,A.A76
****************************************************************************
This structure contains 1-order pseudoknot
o You may want to run DSSR again with the '--nested' option which removes
pseudoknots to get a fully nested secondary structure representation.
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>1ehz nts=76 [whole]
GCGGAUUUAgCUCAGuuGGGAGAGCgCCAGAcUgAAgAPcUGGAGgUCcUGUGtPCGaUCCACAGAAUUCGCACCA
(((((((..((((.....[..)))).((((.........)))).....(((((..]....))))))))))))....
>1ehz-A #1 nts=76 0.09(2.86) [chain] RNA
GCGGAUUUAgCUCAGuuGGGAGAGCgCCAGAcUgAAgAPcUGGAGgUCcUGUGtPCGaUCCACAGAAUUCGCACCA
(((((((..((((.....[..)))).((((.........)))).....(((((..]....))))))))))))....
****************************************************************************
Summary of structural features of 76 nucleotides
Note: the first five columns are: (1) serial number, (2) one-letter
shorthand name, (3) dbn, (4) id string, (5) rmsd (~zero) of base
ring atoms fitted against those in a standard base reference
frame. The sixth (last) column contains a comma-separated list of
features whose meanings are mostly self-explanatory, except for:
turn: angle C1'(i-1)--C1'(i)--C1'(i+1) < 90 degrees
break: no backbone linkage between O3'(i-1) and P(i)
1 G ( A.G1 0.008 anti,~C3'-endo,BI,canonical,non-pair-contact,helix-end,stem-end,coaxial-stack
2 C ( A.C2 0.006 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
3 G ( A.G3 0.021 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
4 G ( A.G4 0.018 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
5 A ( A.A5 0.018 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
6 U ( A.U6 0.011 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
7 U ( A.U7 0.008 anti,~C2'-endo,canonical,non-pair-contact,helix,stem-end,coaxial-stack,junction-loop,phosphate,splayed-apart
8 U . A.U8 0.016 anti,~C3'-endo,non-canonical,non-pair-contact,helix,multiplet,junction-loop,splayed-apart
9 A . A.A9 0.032 anti,~C2'-endo,non-canonical,non-pair-contact,multiplet,junction-loop,phosphate
10 g ( A.2MG10 0.018 modified,turn,anti,~C3'-endo,canonical,non-canonical,non-pair-contact,helix,stem-end,coaxial-stack,multiplet,junction-loop
11 C ( A.C11 0.005 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
12 U ( A.U12 0.012 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack,multiplet
13 C ( A.C13 0.013 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,multiplet,hairpin-loop,kissing-loop
14 A . A.A14 0.025 anti,~C3'-endo,non-canonical,non-pair-contact,helix,multiplet,hairpin-loop,kissing-loop,phosphate
15 G . A.G15 0.021 anti,~C3'-endo,non-canonical,non-pair-contact,helix,hairpin-loop,kissing-loop
16 u . A.H2U16 0.188 modified,anti,~C3'-endo,non-canonical,non-pair-contact,helix-end,hairpin-loop,kissing-loop,cap-donor,cap-acceptor,phosphate,splayed-apart
17 u . A.H2U17 0.201 modified,turn,anti,~C3'-endo,non-stack,non-pair-contact,hairpin-loop,kissing-loop,splayed-apart
18 G . A.G18 0.028 anti,~C2'-endo,BII,non-canonical,non-pair-contact,helix,hairpin-loop,kissing-loop,phosphate,splayed-apart
19 G [ A.G19 0.026 pseudoknotted,anti,~C2'-endo,isolated-canonical,non-pair-contact,helix-end,hairpin-loop,kissing-loop,phosphate,splayed-apart
20 G . A.G20 0.018 anti,~C3'-endo,non-stack,non-pair-contact,hairpin-loop,kissing-loop,phosphate,splayed-apart
21 A . A.A21 0.018 anti,~C3'-endo,BI,non-canonical,non-pair-contact,multiplet,hairpin-loop,kissing-loop,phosphate
22 G ) A.G22 0.017 anti,~C3'-endo,BI,canonical,non-canonical,non-pair-contact,helix,stem-end,coaxial-stack,multiplet,hairpin-loop,kissing-loop
23 A ) A.A23 0.030 anti,~C3'-endo,BI,canonical,non-canonical,non-pair-contact,helix,stem,coaxial-stack,multiplet,phosphate
24 G ) A.G24 0.013 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
25 C ) A.C25 0.011 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,multiplet,junction-loop
26 g . A.M2G26 0.013 modified,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,junction-loop
27 C ( A.C27 0.011 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,junction-loop
28 C ( A.C28 0.007 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
29 A ( A.A29 0.019 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
30 G ( A.G30 0.013 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,hairpin-loop
31 A . A.A31 0.007 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,hairpin-loop
32 c . A.OMC32 0.008 modified,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,hairpin-loop
33 U . A.U33 0.014 u-turn,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix-end,hairpin-loop,cap-acceptor
34 g . A.OMG34 0.021 modified,turn,u-turn,anti,~C3'-endo,BI,non-pair-contact,hairpin-loop
35 A . A.A35 0.006 u-turn,anti,~C3'-endo,BI,non-pair-contact,hairpin-loop,cap-donor,phosphate
36 A . A.A36 0.015 u-turn,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix-end,hairpin-loop,phosphate
37 g . A.YYG37 0.015 modified,anti,~C3'-endo,BI,non-pair-contact,hairpin-loop,phosphate
38 A . A.A38 0.008 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,hairpin-loop
39 P . A.PSU39 0.004 modified,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,hairpin-loop
40 c ) A.5MC40 0.005 modified,anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,hairpin-loop
41 U ) A.U41 0.013 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
42 G ) A.G42 0.010 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
43 G ) A.G43 0.025 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,junction-loop
44 A . A.A44 0.015 anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,junction-loop
45 G . A.G45 0.026 anti,~C3'-endo,BI,non-canonical,non-pair-contact,multiplet,junction-loop
46 g . A.7MG46 0.022 modified,anti,~C2'-endo,non-canonical,non-pair-contact,multiplet,junction-loop,splayed-apart
47 U . A.U47 0.007 turn,anti,~C2'-endo,non-stack,non-pair-contact,junction-loop,splayed-apart
48 C . A.C48 0.015 turn,anti,~C2'-endo,non-canonical,non-pair-contact,helix,junction-loop,cap-acceptor,phosphate,splayed-apart
49 c ( A.5MC49 0.020 modified,anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,junction-loop,phosphate,splayed-apart
50 U ( A.U50 0.024 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack,phosphate
51 G ( A.G51 0.030 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
52 U ( A.U52 0.020 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
53 G ( A.G53 0.027 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,hairpin-loop,kissing-loop
54 t . A.5MU54 0.010 modified,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,hairpin-loop,kissing-loop
55 P . A.PSU55 0.024 modified,u-turn,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix,hairpin-loop,kissing-loop,cap-acceptor
56 C ] A.C56 0.014 pseudoknotted,turn,u-turn,anti,~C3'-endo,BI,isolated-canonical,non-pair-contact,helix-end,hairpin-loop,kissing-loop
57 G . A.G57 0.017 u-turn,anti,~C3'-endo,BI,non-pair-contact,hairpin-loop,kissing-loop,cap-donor,phosphate
58 a . A.1MA58 0.012 modified,u-turn,anti,~C2'-endo,BII,non-canonical,non-pair-contact,helix,hairpin-loop,kissing-loop,phosphate
59 U . A.U59 0.029 turn,anti,~C3'-endo,BI,non-canonical,non-pair-contact,helix-end,hairpin-loop,kissing-loop,cap-donor
60 C . A.C60 0.016 anti,~C2'-endo,non-pair-contact,hairpin-loop,kissing-loop,phosphate,splayed-apart
61 C ) A.C61 0.012 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,hairpin-loop,kissing-loop,splayed-apart
62 A ) A.A62 0.016 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
63 C ) A.C63 0.016 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
64 A ) A.A64 0.014 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
65 G ) A.G65 0.017 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,junction-loop
66 A ) A.A66 0.016 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem-end,coaxial-stack,junction-loop
67 A ) A.A67 0.020 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
68 U ) A.U68 0.011 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
69 U ) A.U69 0.015 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
70 C ) A.C70 0.018 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
71 G ) A.G71 0.013 anti,~C3'-endo,BI,canonical,non-pair-contact,helix,stem,coaxial-stack
72 C ) A.C72 0.006 anti,~C3'-endo,BI,canonical,non-pair-contact,helix-end,stem-end,coaxial-stack
73 A . A.A73 0.010 anti,~C3'-endo,BI,non-pair-contact,ss-non-loop
74 C . A.C74 0.007 anti,~C3'-endo,BI,non-pair-contact,ss-non-loop
75 C . A.C75 0.007 anti,~C3'-endo,BII,non-pair-contact,ss-non-loop,splayed-apart
76 A . A.A76 0.010 anti,~C2'-endo,non-stack,non-pair-contact,ss-non-loop,splayed-apart
****************************************************************************
List of 14 additional files
1 dssr-pairs.pdb -- an ensemble of base pairs
2 dssr-multiplets.pdb -- an ensemble of multiplets
3 dssr-stems.pdb -- an ensemble of stems
4 dssr-helices.pdb -- an ensemble of helices (coaxial stacking)
5 dssr-hairpins.pdb -- an ensemble of hairpin loops
6 dssr-junctions.pdb -- an ensemble of junctions (multi-branch)
7 dssr-2ndstrs.bpseq -- secondary structure in bpseq format
8 dssr-2ndstrs.ct -- secondary structure in connectivity table format
9 dssr-2ndstrs.dbn -- secondary structure in dot-bracket notation
10 dssr-torsions.txt -- backbone torsion angles and suite names
11 dssr-splays.pdb -- an ensemble of splayed-apart units
12 dssr-Uturns.pdb -- an ensemble of U-turn motifs
13 dssr-stacks.pdb -- an ensemble of stacks
14 dssr-atom2bases.pdb -- an ensemble of atom-base stacking interactions
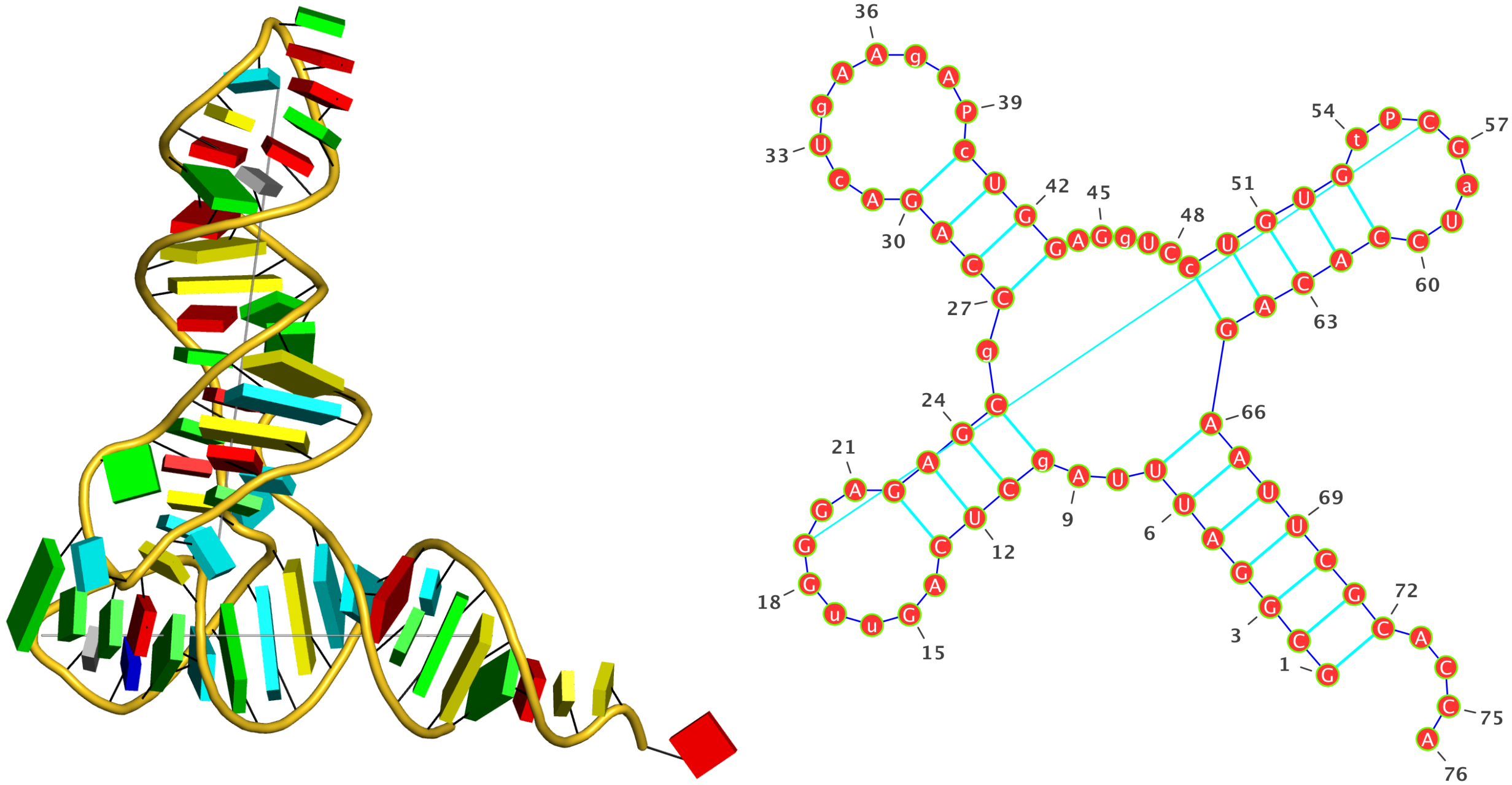
Note: shown above, the 3-dimensional schematic images (with rectangular base blocks) were created with the 3DNA
blocview program to generate
.r3d-formatted files that were ray-traced using PyMOL. The 2-dimensional diagrams were produced with
VARNA: Visualization Applet for RNA using DSSR-derived base sequence and dot-bracket notation of secondary structure:
>1msy-A #1 RNA with 27 nts
UGCUCCUAGUACGUAAGGACCGGAGUG
.(((((.....(....)....))))).
>1ehz-A #1 RNA with 76 nts
GCGGAUUUAgCUCAGuuGGGAGAGCgCCAGAcUgAAgAPcUGGAGgUCcUGUGuPCGaUCCACAGAAUUCGCACCA
(((((((..((((.....[..)))).((((.........)))).....(((((..]....))))))))))))....


