The article "
Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures" has been published in the 2019 web server issue of
Nucleic Acids Research (NAR), and
highlighted in the cover page! Co-authored by Shuxiang Li, Wilma Olson and me, this w3DNA 2.0 paper is a significant contribution to the field of nucleic acids structures, and it will undoubtedly push the popularity of 3DNA to a new level.
At nine pages, the paper is not a typical NAR web-server publication but contains several new parameters introduced in 3DNA after the initial publication of
w3DNA in 2009. The abstract is shown below:
Web 3DNA (w3DNA) 2.0 is a significantly enhanced version of the widely used w3DNA server for the analysis, visualization, and modeling of 3D nucleic-acid-containing structures. Since its initial release in 2009, the w3DNA server has continuously served the community by making commonly-used features of the 3DNA suite of command-line programs readily accessible. However, due to the lack of updates, w3DNA has clearly shown its age in terms of modern web technologies and it has long lagged behind further developments of 3DNA per se. The w3DNA 2.0 server presented here overcomes all known shortcomings of w3DNA while maintaining its battle-tested characteristics. Technically, w3DNA 2.0 implements a simple and intuitive interface (with sensible defaults) for increased usability, and it complies with HTML5 web standards for broad accessibility. Featurewise, w3DNA 2.0 employs the most recent version of 3DNA, enhanced with many new functionalities, including: the automatic handling of modified nucleotides; a set of ‘simple’ base-pair and step parameters for qualitative characterization of non-Watson–Crick double- helical structures; new structural parameters that integrate the rigid base plane and the backbone phosphate group, the two nucleic acid components most reliably determined with X-ray crystallography; in silico base mutations that preserve the backbone geometry; and a notably improved module for building models of single-stranded RNA, double- helical DNA, Pauling triplex, G-quadruplex, or DNA structures ‘decorated’ with proteins. The w3DNA 2.0 server is freely available, without registration, at http://web.x3dna.org.
This section is dedicated to topics on reproducing the results reported in the w3DNA 2.0 article, with scripts and related data files. We welcome any questions and comments you may have, here on the 3DNA Forum.
Cover imageCaption: Examples of customized molecular models that can be generated with 3DNA: (top) a chromatin-like, nucleosome-decorated DNA with the structures of known histone-DNA assemblies placed at user-defined binding sites; (lower left) molecular schematic of a DNA trinucleotide diphosphate illustrating the base planes and reference frames used to construct and analyze 3D nucleic acid-containing structures; (lower right) customized single-stranded tRNA model built from a user-defined base sequence and a set of rigid-body parameters describing the desired placement of successive bases. Color code of base blocks: A, red; C, yellow; G, green; T, blue; U, cyan.
Figure 1. Summary of web 3DNA 2.0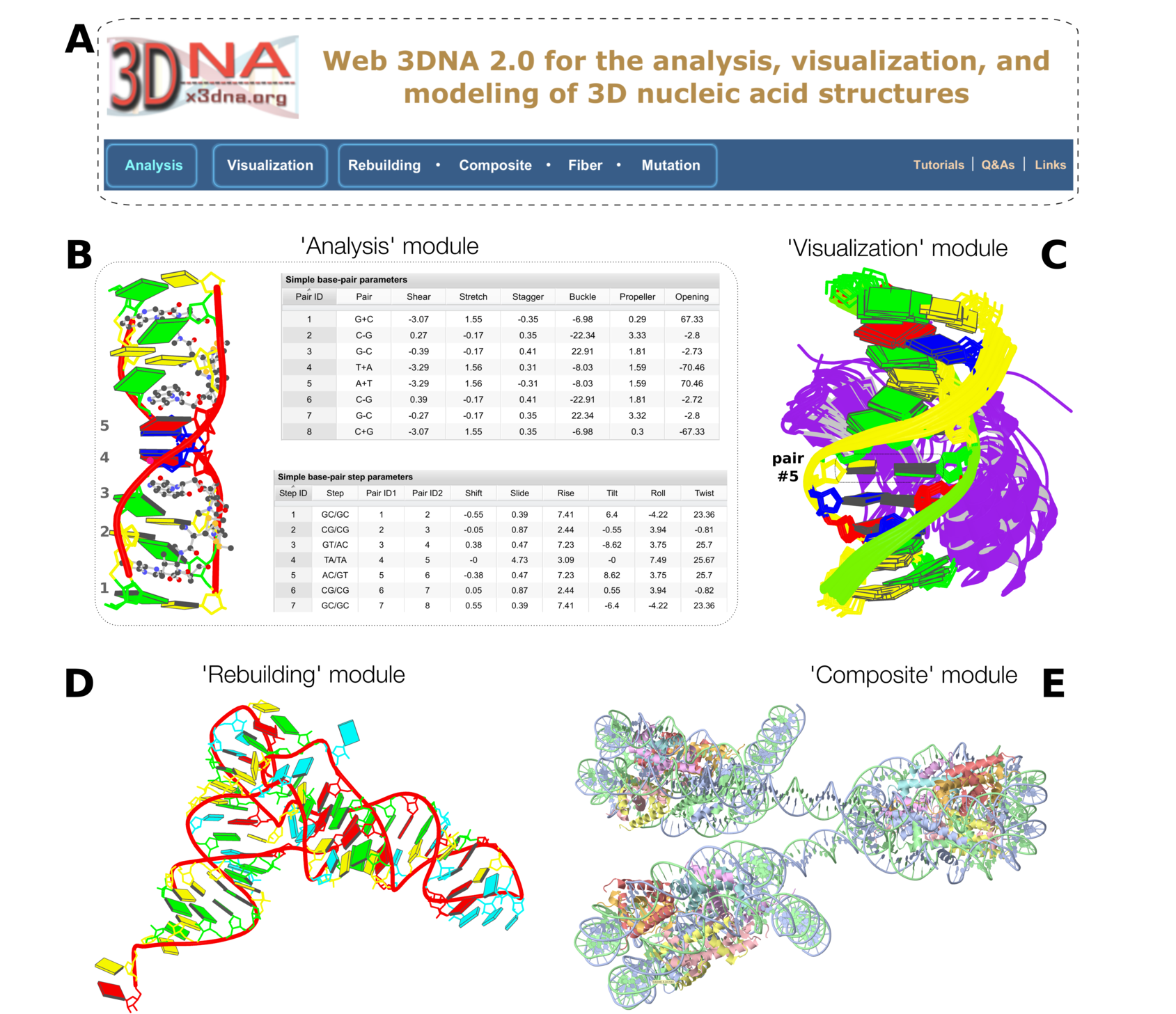
Figure 1. Summary of web 3DNA 2.0. (A) The homepage, highlighting the three major components of the server (boxed) and links to key resources. (B) Excerpt from the ‘Analysis’ of a drug–DNA complex (PDB entry 1xvk) (38), showing a base-block image and tabulations of the ‘simple’ base-pair and step parameters. (C) Schematic ‘Visualization’ of an ensemble of NMR structures of a protein–DNA complex (PDB entry 2moe) (31). The models are aligned locally in the reference frame of the fifth base pair, with its minor-groove edge (colored black) facing the viewer. The protein, colored purple, binds in the major groove of the DNA. (D) An example of ‘Rebuilding’ a model from the local base-pair and step parameters obtained from an ‘Analysis’ of a tRNA structure (PDB entry: 1fir) (33). (E) ‘Composite’ model of a DNA ‘decorated’ with proteins. Here a nucleosome (PDB entry: 4xzq) (34) is used as a template to construct a three-nucleosome, chromatin-like structure. Color code for base rectangular blocks: A, red; C, yellow; G, green; T, blue; U, cyan. (B–D) were generated automatically via the 3DNA ‘blocview’ program, which calls MolScript (24) and Raster3D (25); (E) was rendered using JSmol (22). The annotations were created using Inkscape (
https://inkscape.org).
Figure 2. New structural parameters that connect base and backbone atoms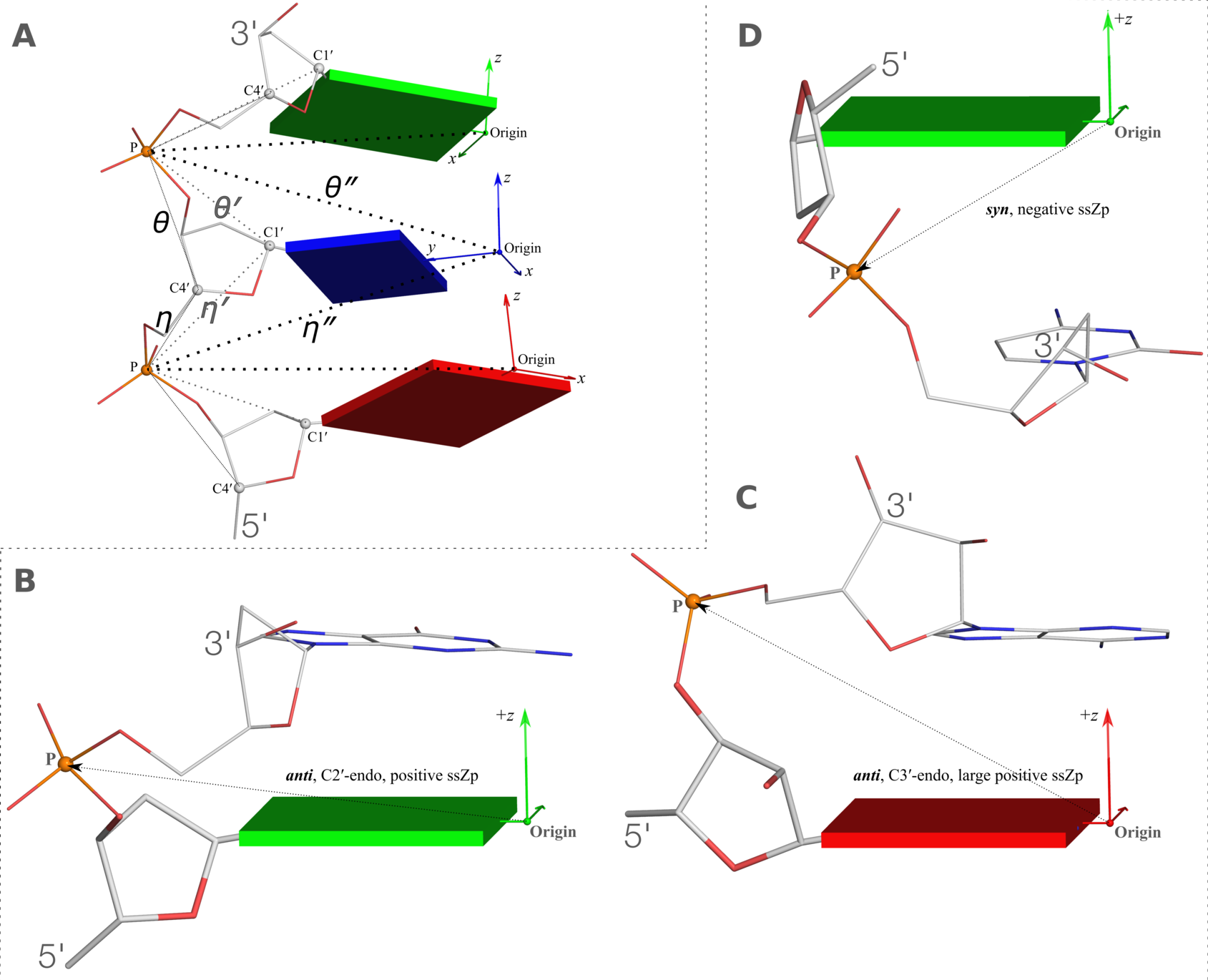
Figure 2. New structural parameters that connect base and backbone atoms. (A) A pair of virtual torsion angles (η′′ and θ′′) that are based on the positions of the phosphorus atoms (P) and the origins of the intervening bases in their standard base reference frames (7). Two closely related forms of virtual backbone torsion angles are depicted for comparison: the classic version (η and θ) defined by the P and C4′ atoms (27), and a more recent variant (η′ and θ′) based on the P and C1′ atoms (28). Here, an ApTpG trinucleotide from a B-DNA fiber model is used for illustration, with base reference frames attached. Note that the standard base frames of purines and pyrimidines are symmetrical with respect to the dyad of an idealized Watson–Crick base pair, and thus independent of base identity (21): the base origin is accordingly more displaced from the atoms of T (a pyrimidine) than those of A or G (purines). (B–D) Single-stranded phosphate displacement, ssZp, in representative helical structures showing: (B) a small, positive number for a GpG step from a B-DNA fiber model where a C2′-endo sugar attaches to a base in an anti conformation (ssZp = +1.84 Å); (C) a large, positive value for an ApA step from an RNA fiber model where a C3′-endo sugar attaches to a base in an anti conformation (ssZp = +4.38 Å); (D) a negative value for a GpC step from a Z-DNA fiber model where the G adopts a syn conformation (ssZp = −1.74 Å). Color code for base blocks: A, red; G, green; T, blue. The illustrations were generated with DSSR (21) and PyMOL (
https://pymol.org).
Figure 3. Commonly-used fiber models and in silico base mutations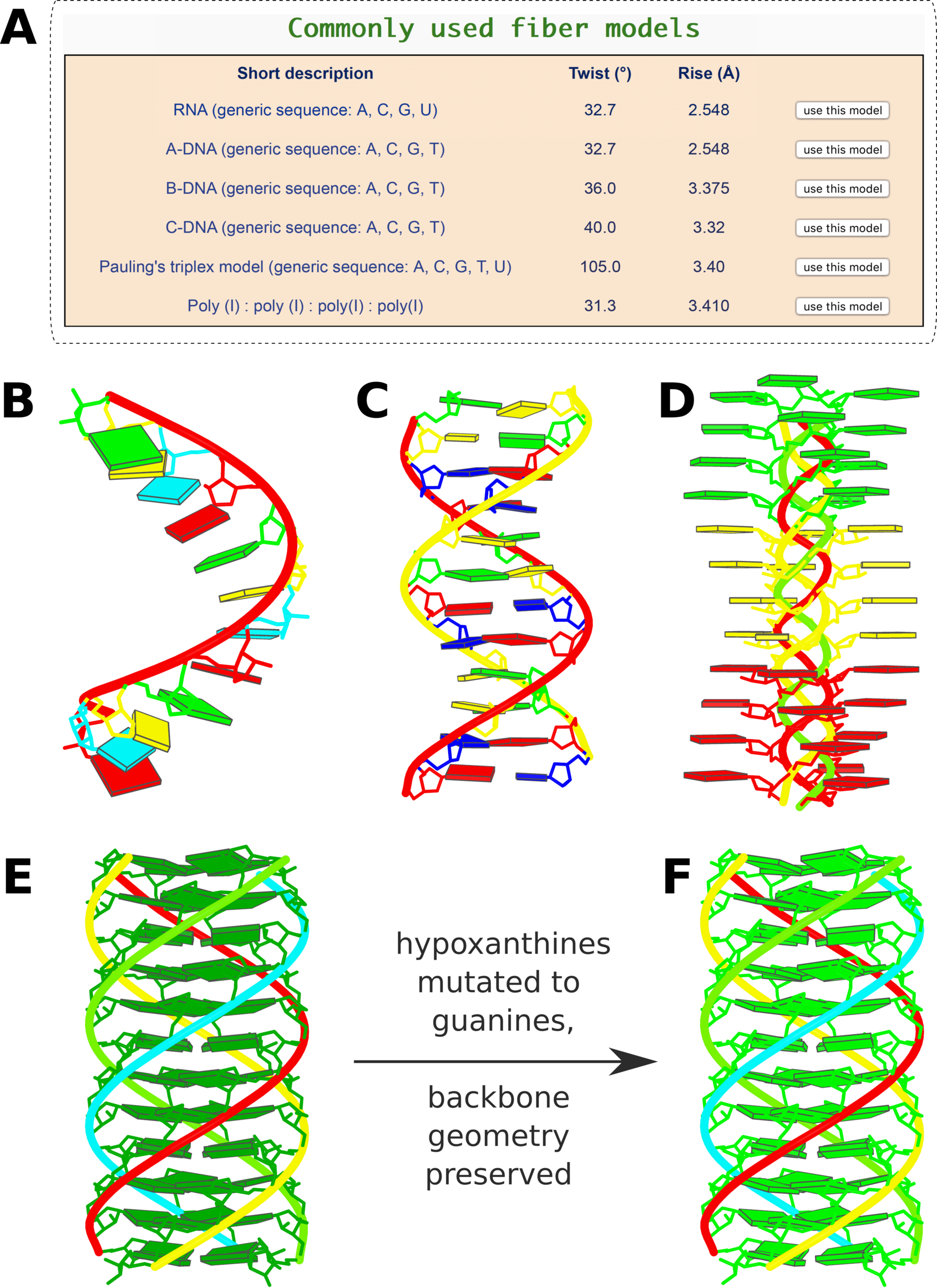
Figure 3. Commonly-used fiber models and in silico base mutations. (A) Six commonly used models highlighted in the ‘Fiber’ module: single-stranded RNA, double-helical A-, B-, and C-form DNA, the Pauling triplex model (32), and the parallel polyI:polyI:polyI:polyI quadruplex. (B) Single-stranded RNA fiber model of base sequence AUCGAUCGAUCG. (C) Double-helical B-DNA fiber model with sequence ATCGATC- GATCG on the leading strand. (D) Pauling triplex model with each strand of sequence AAAACCCCGGGG. (E) parallel polyI:polyI:polyI:polyI quadruplex model with 12 layers of hydrogen-bonded hypoxanthine tetrads. Models in (B-E) were generated using the default settings on the w3DNA 2.0 server, each taking just two mouse clicks. (F) All hypoxanthine bases along the poly I chains mutated to guanine via the ‘Mutation’ module, leading to a parallel G-quadruplex. Color code for base blocks: A, red; C, yellow; G, green; T, blue; U, cyan; I, dark green.
Supplementary Data (w3DNA2.0-supp.pdf), serving as a manual to w3DNA 2.0. Note that the supplementary PDF here contains
DOI links to the w3DNA 2.0 publication.
Graphical abstract (taken from Fig. 1E):


