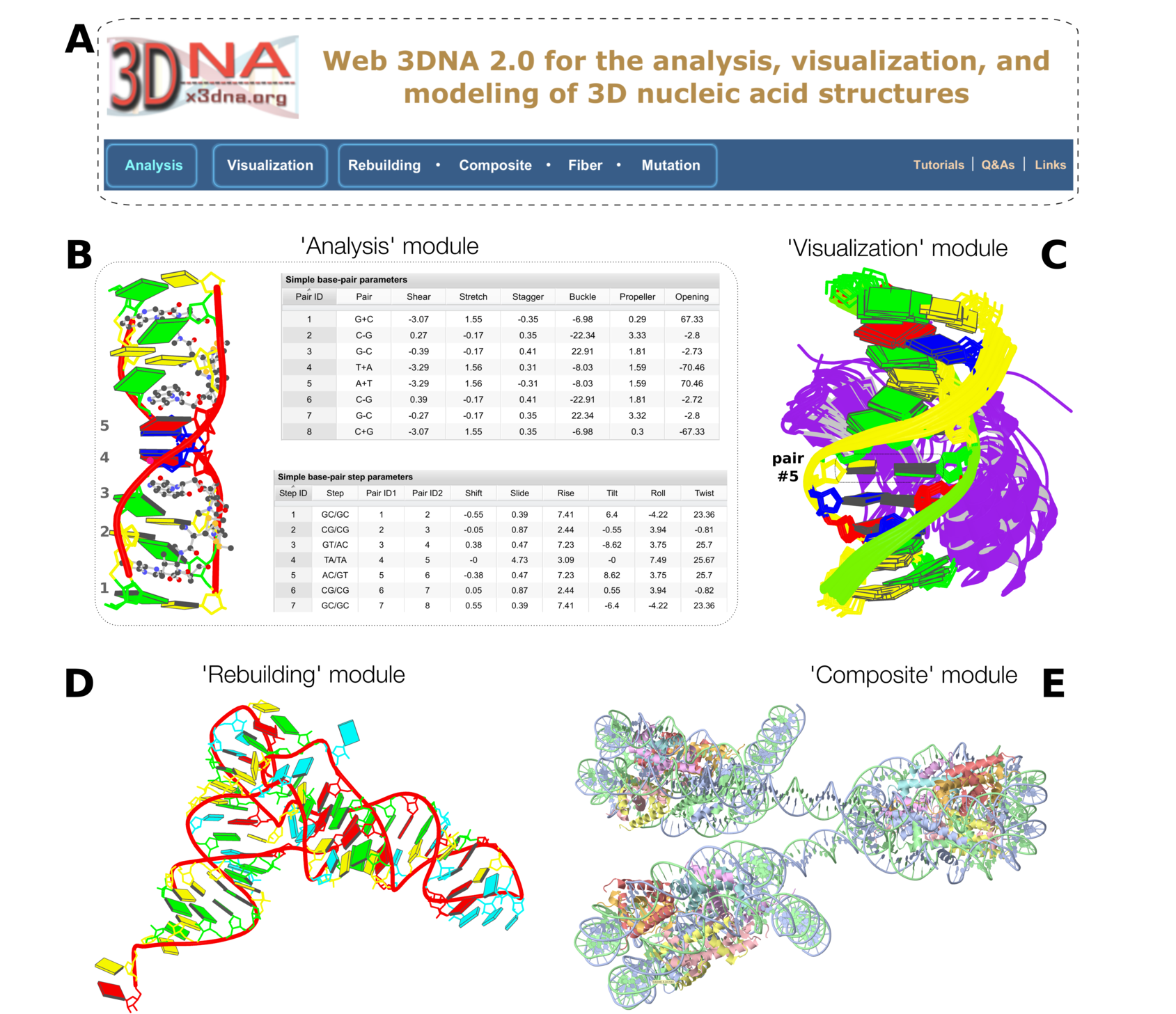
Figure 1. Summary of web 3DNA 2.0. (A) The homepage, highlighting the three major components of the server (boxed) and links to key resources. (B) Excerpt from the ‘Analysis’ of a drug–DNA complex (PDB entry 1xvk) (38), showing a base-block image and tabulations of the ‘simple’ base-pair and step parameters. (C) Schematic ‘Visualization’ of an ensemble of NMR structures of a protein–DNA complex (PDB entry 2moe) (31). The models are aligned locally in the reference frame of the fifth base pair, with its minor-groove edge (colored black) facing the viewer. The protein, colored purple, binds in the major groove of the DNA. (D) An example of ‘Rebuilding’ a model from the local base-pair and step parameters obtained from an ‘Analysis’ of a tRNA structure (PDB entry: 1fir) (33). (E) ‘Composite’ model of a DNA ‘decorated’ with proteins. Here a nucleosome (PDB entry: 4xzq) (34) is used as a template to construct a three-nucleosome, chromatin-like structure. Color code for base rectangular blocks: A, red; C, yellow; G, green; T, blue; U, cyan. (B–D) were generated automatically via the 3DNA ‘blocview’ program, which calls MolScript (24) and Raster3D (25); (E) was rendered using JSmol (22). The annotations were created using Inkscape (https://inkscape.org).
Fig. 1A is simply a screenshot of the homepage header, including the six modules on the left and three related links on the right.
Fig. 1B is about the 'Analysis' module using
PDB entry 1xvk as an example. The block image corresponds to the left view and the two tables ("Simple base-pair parameters" and "Simple base-pair step parameters") are screenshots, all from the w3DNA 2.0 output. The new 'simple' parameters have been devised for a more intuitive,
qualitative characterization of non-Watson-Crick base pairs. The PDB entry 1xvk was chosen because of the thread "
zero or negative helical rise?" on the Forum started on March 25, 2013 (and "
question on +/- signs of local bp parameters" on January 11, 2013).
See also:
Fig. 1C illustrates the 'Visualization' module using the NMR ensemble from
PDB entry 2moe as an example. Among the 20 models in 2moe, the first 10 models are used here by default. The image can be generated directly using the default settings.
Fig. 1D showcases the 'Rebuilding' module with a customized base sequence and corresponding rigid-body parameters, using a tRNA structure based on
PDB entry 1fir as an example. Simply click the buttons with default settings will lead to the image shown here. See also section "S2.2 Rebuilding a single-stranded RNA structure" in the
supplemental PDF.
Fig. 1E exhibits the 'Composite' module to create a chromatin-like structure with three nucleosomes. The tutorial "
Example 4-2: Construction of a chromatin-like model" on the website (and in the
supplemental PDF) provides the detailed steps.
The image shown below was interactively rotated in JSmol from the direct output view of w3DNA 2.0, and then rendered via the console, using the following command (as the one used in the
DSSR-Jmol paper):
write PNGJ 3000 3000 w3DNA2.0-fig1E-jmol.png 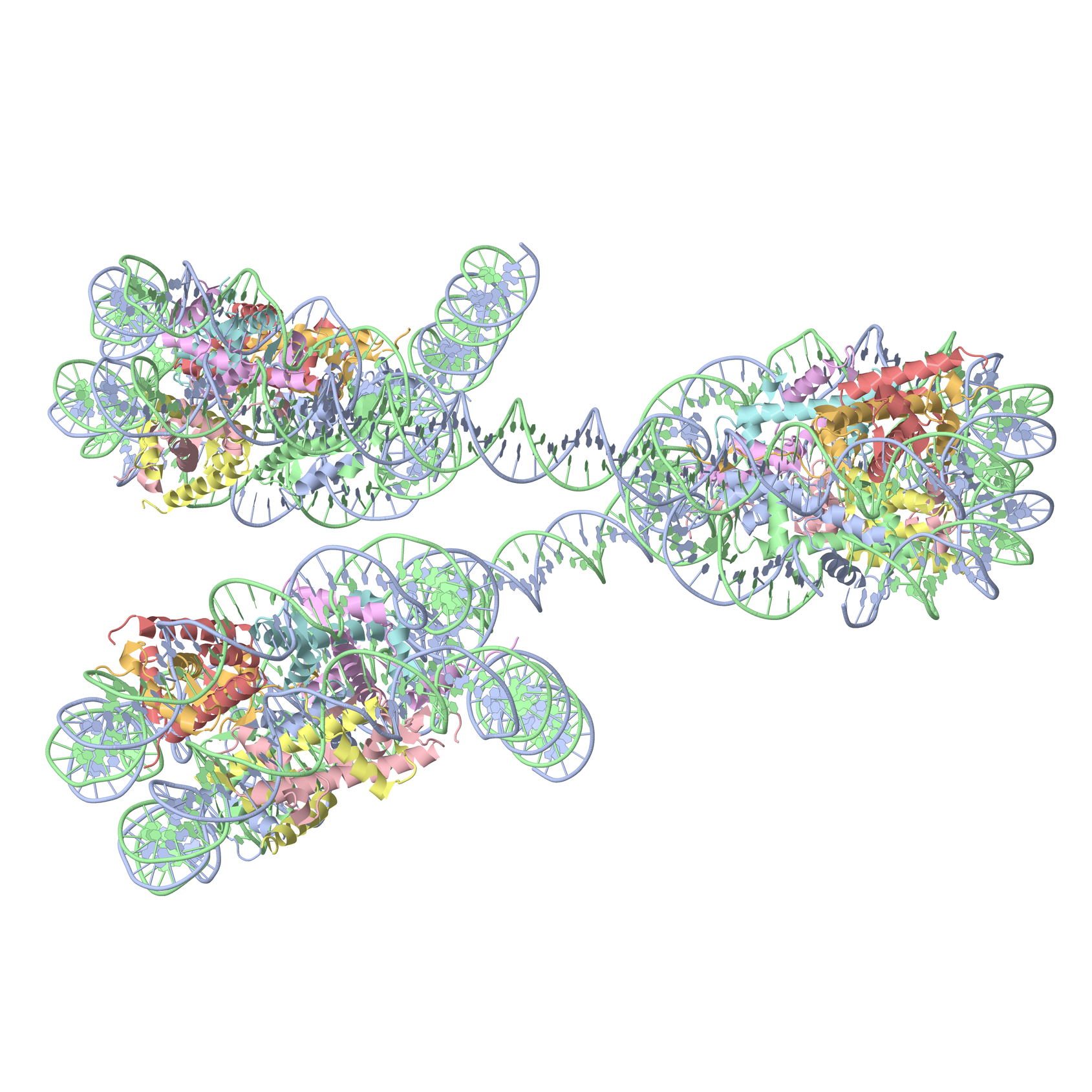
It is worth noting that PNGJ option retains the atomic coordinates used to create the PNG image. As a result, the file
w3DNA2.0-fig1E-jmol.png can be reloaded to Jmol/JSmol for interactive manipulation.
Fig. 1D and 1E use
Phenix to perform a the
base-restraint optimization of backbone geometry. See also the FAQ entry "
How do I build nucleic acid structures with sugar-phosphate backbone?"


