Hi Xiang-Jun and other 3DNA users,
It's very nice to meet you, thank your for your time checking this topic. I am a masters student currently working with Molecular Dynamics trajectories of DNA using NAMD and I look forward to use the X3DNA Ensemble in my research.
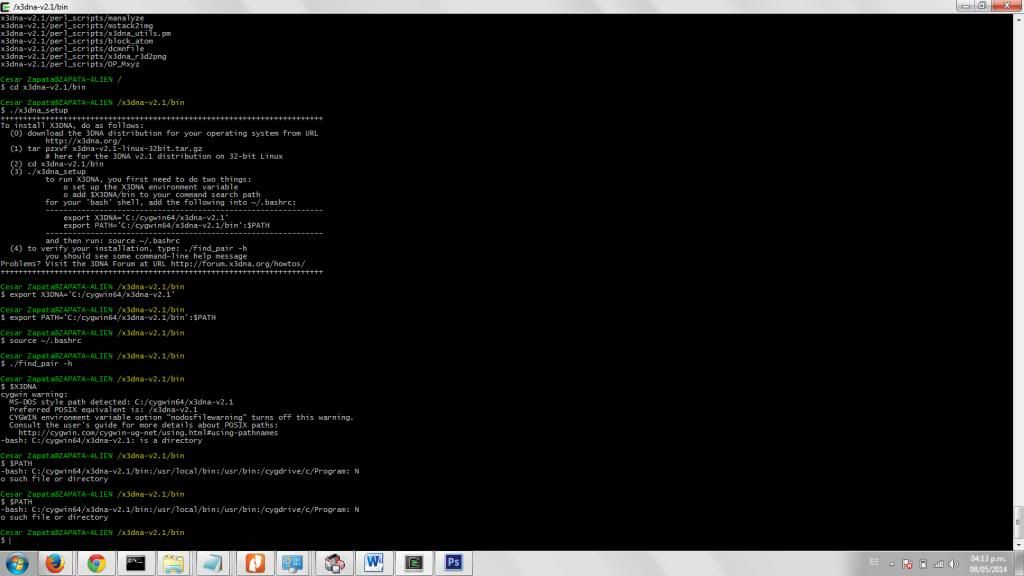
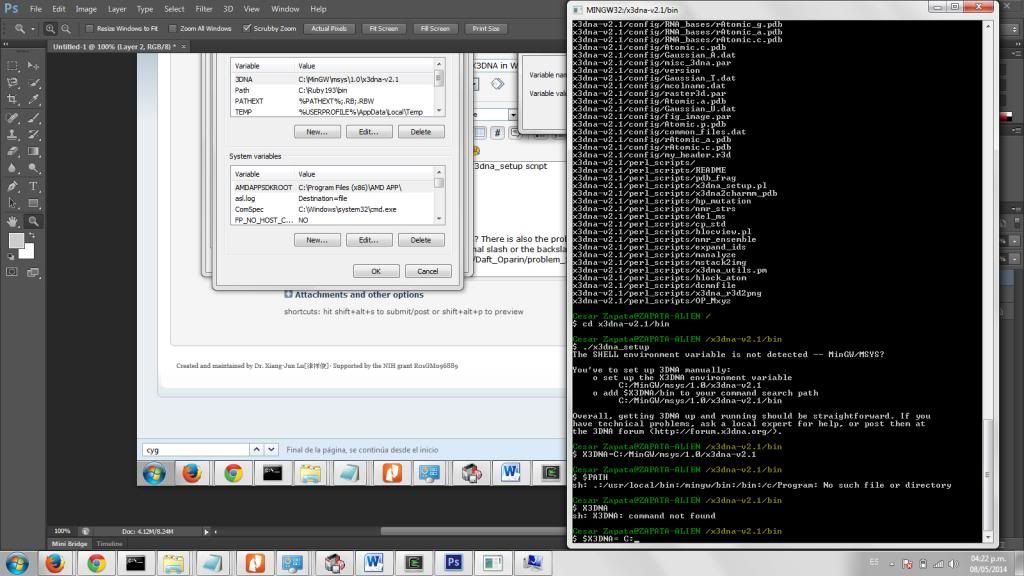
I am currently trying to install X3DNA in my Windows 7-64 bits-based computer. I have already installed Ruby, and have downloaded both options of Cygwin and MinGW-MSYS. I can't finish the installation. Both programs seem to run and never give me any error message, but when I try the ./find_pairs -h it doesn't do anything.
I don't think have any problems until running the ./x3dna_setup script, setting up the variables and running that source ~/.bashrc which I can't seem to find.
It says:
(3) ./x3dna_setup
to run X3DNA, you first need to do two things:
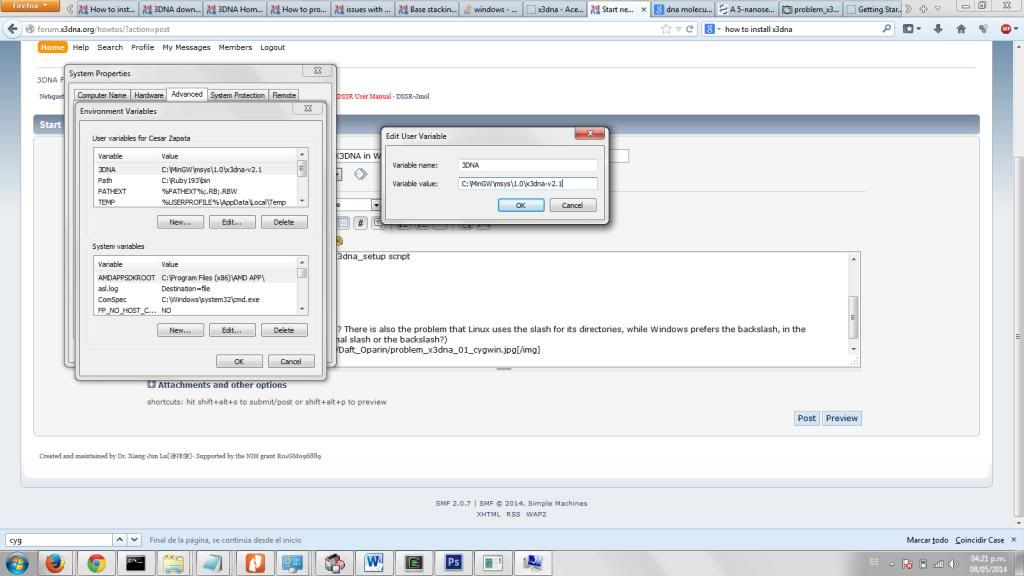
* set up the X3DNA environment variable
(Are we talking about the variable in Windows? There is also the problem that Linux uses the slash for its directories, while Windows prefers the backslash, in the Windows User Variables should I use the normal slash or the backslash?)
Or perhaps we are talking about the internal variables of the Unix-like shells?
Once the program is installed, will it be able to be called from the normal Windows command line or will it always have to be called through Cygwin or MinGW?
I seem to be lacking a crucial part of the process.
Any help would be greatly appreciated, and thanks a lot again.


