See
DSSR --
a new component in 3DNA for Defining the (Secondary) Structures of RNA, and more... note added on Saturday, March 16, 2013.
While 3DNA stands for
3d-NA instead of 3-
DNA, my experience tells me that there are noticeable confusions in the structural bioinformatics community as to its capabilities for RNA structures. The favicon and logo of the new 3DNA homepage and forum have been designed to help clarify the issue. To make the message even clearer, I have set up this specific section titled "RNA structures".
This starting post summarizes some of 3DNA's facilities for the analysis and modeling of RNA structures, using concrete examples. Note PDB entry
6tna (NDB id
trna04) is for the crystal structure of yeast phenylalanine transfer RNA. All the data files and images are generated directly from the commands given, and
the results should be exactly reproducible.
- Generate regular double-stranded or single-stranded RNA models of arbitrary sequence:
fiber -r -seq=accugggga dsRNA.pdb
fiber -r -seq=accugggga -s ssRNA.pdb
As of v2.1, fiber has three new options: -r for RNA, -seq for specifying sequence (of lower, UPPER or Mixed case) directly on the command-line, and -s for single-stranded structure. Download ssRNA.pdb of sequence accugggga.
- Calculate all backbone torsion angles:
analyze -tor=6tna.tor 6tna.pdb # as of 3DNA v2.1
find_pair -s 6tna.pdb stdout | analyze stdin
Note the -s option; as of v2.1, explicit input file name (here stdin) is required for analyze. The output file is saved in file 6tna.outs.
- Detect coaxially stacked helices:
find_pair 6tna.pdb 6tna.inp
ex_str -1 hel_regions.pdb h1.pdb
ex_str -2 hel_regions.pdb h2.pdb
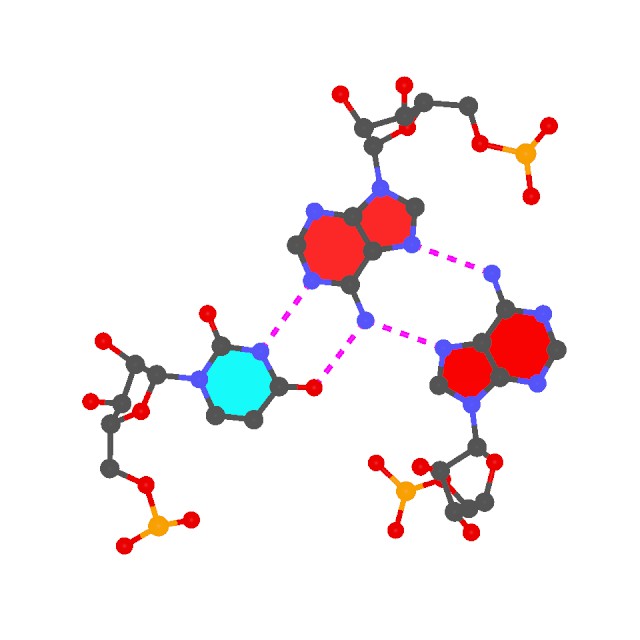
The two helical regions, saved in file h1.pdb and h2.pdb correspond to the two arms of the L-shaped tRNA structure (see the 6tna.jpg image below). - Find all (non-canonical) base-pairs, and higher-order base associations. Here only one of the three base triplets is shown:
find_pair -p 6tna.pdb 6tna.mbp
ex_str -1 multiplets.pdb triplet1.pdb
r3d_atom -od -r=0.1 -b=0.2 triplet1.pdb stdout | render -jpeg > triplet1.jpg
Note the -p option. Higher-order base associations (i.e., with 3+ bases) are saved in file multiplets.pdb where each multiplet is automatically set in the most extended views to the mean plane of the bases. Here render from Raster3D is used to convert 3DNA-generated .r3d file into a jpeg image.
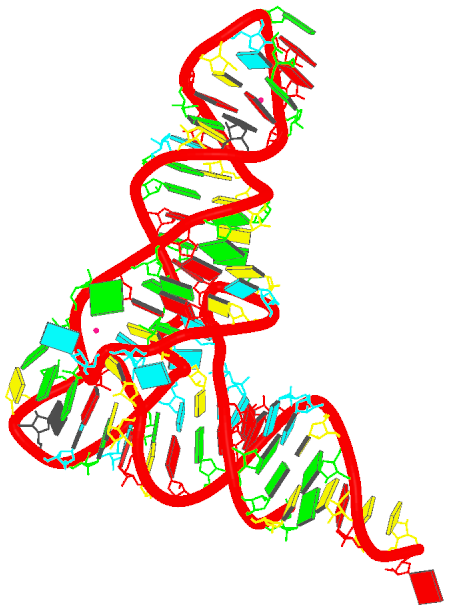
- Generate the schematics base block image in the overall "best" view:
blocview -i=6tna.jpg 6tna.pdb
The image is named 6tna.jpg, and is shown below:
In my understanding, 3DNA's RNA functionalities outlined above are distinct from the well-known
FR3D program developed by the Leontis-Zirbel team. 3DNA and FR3D are somewhat complementary in purpose and running style. As a specific example, as noted in the
2003 3DNA NAR paper excerpted below, 3DNA has unique features for base-pair classification that are complementary to the 3-edge (Watson-Crick edge, Hoogsteen edge and sugar edge) based Leontif-Westhof scheme.
Since the six base pair parameters uniquely define the relative position and orientation of two bases, they can be used to reconstruct the base pair. Moreover, the parameters provide a simple mechanism for classification of structures (55) and database searching (X.-J. Lu, Y. Xin and W.K. Olson, unpublished data). Among the six base pair parameters, only Shear, Stretch and Opening are critical in characterizing key hydrogen bonding features, i.e. base pair type: Shear and Stretch define the relative offset of the two base origins in the mean base pair plane and Opening is the angle between the two x-axes with respect to the average normal to the base pair plane (see upper left panel in Fig. 1). For the Hoogsteen A+U base pair shown in Figure 2b, Shear is 0.5 Å , Stretch –3.5 Å and Opening 70°. Buckle, Propeller and Stagger, in contrast, are secondary parameters, which simply describe the imperfections, i.e. non-planarity, of a given base pair.
…………………………………………………………………………………………………………………………………………
Further details of the base pair search algorithm and the composition and geometries of all base pairing interactions observed to date in well-resolved RNA structures will be reported elsewhere (X.-J. Lu, Y. Xin and W.K. Olson, manuscript in preparation).
I am interested in extending 3DNA further into the RNA (structural) world. If you would like to collaborate on some specific project in this broad and important area, please drop me a message. Now that this section is open, I am hoping to see more applications of 3DNA in RNA structures. Questions, opinions, or comments? Please do not hesitate to post them here!
See also:
Xiang-Jun


