1151
RNA structures (DSSR) / Re: DSSR - List of bases involved in hairpins?
« on: March 14, 2013, 03:29:27 pm »
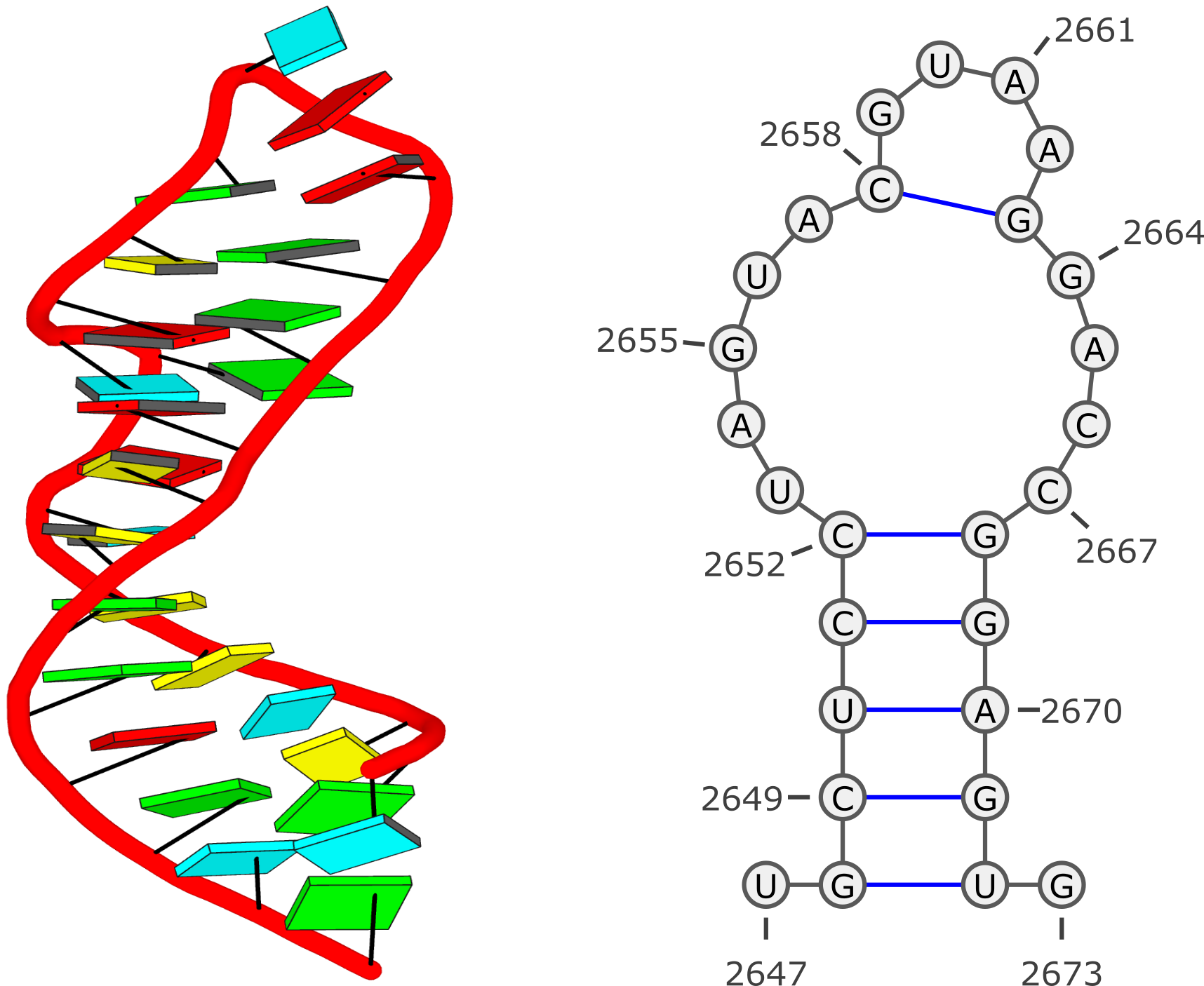
I've just updated DSSR to beta-r04-on-20130314. Among other things, now all nucleotides in hairpin loops are explicitly listed in addition to information reported before. So for 1msy, the new output looks like below:
I have some reasons to report hairpin loops differently from other types of loops (bulges, internal loops, junctions), one being to follow the convention. For example, at first glance, one would immediately see that 1msy contains a GUAA tetra-loop (of the most common GNRA type).
I am not aware of a consistent way to name other loops in the literature of RNA structures, so I've come up with my own convention. I'd like to hear what the community has to say when DSSR gains more popularity in the RNA structural world, and make adjustments accordingly.
HTH,
Xiang-Jun
Code: [Select]
List of 1 hairpin loop(s)
1 nts=4 GUAA closed by pair {A.C2658+A.G2663 [CG], #-1}
A.C2658+A.G2659+A.U2660+A.A2661+A.A2662+A.G2663 [CGUAAG]I have some reasons to report hairpin loops differently from other types of loops (bulges, internal loops, junctions), one being to follow the convention. For example, at first glance, one would immediately see that 1msy contains a GUAA tetra-loop (of the most common GNRA type).
I am not aware of a consistent way to name other loops in the literature of RNA structures, so I've come up with my own convention. I'd like to hear what the community has to say when DSSR gains more popularity in the RNA structural world, and make adjustments accordingly.
HTH,
Xiang-Jun


 . I'll get them fixed shortly -- hopefully by tomorrow. With the next release, the program would be quite robust, I believe.
. I'll get them fixed shortly -- hopefully by tomorrow. With the next release, the program would be quite robust, I believe.