Hi Prasun Kumar,
Thanks for the 'bug report'. To facilitate our communications, I am putting your attached file in pre-formatted form (content of '
1BNA_groove_width.txt').
3DNA
Minor Groove Major Groove
P-P Refined P-P Refined
1 CG/CG --- --- --- ---
2 GC/GC --- --- --- ---
3 CG/CG 13.8 --- 17.3 ---
4 GA/TC 12.0 12.0 17.3 17.3
5 AA/TT 10.5 10.5 17.5 17.3
6 AT/AT 9.9 9.9 16.8 16.3
7 TT/AA 9.3 9.3 17.4 17.4
8 TC/GA 10.0 9.9 18.5 18.4
9 CG/CG 11.0 --- 17.8 ---
10 GC/GC --- --- --- ---
11 CG/CG --- --- --- ---
NUPARM
Inter-chain P--P distances
Strand2: 5'-->3'
Strand1:
PP 2. P 37.07 33.00 27.61 22.52 19.58 17.41 17.32 18.27 18.81 18.50 17.96
PP 3. P 33.23 28.47 22.91 18.57 17.60 17.32 18.20 18.97 19.06 18.14 16.78
PP 4. P 28.98 24.01 18.93 16.44 17.53 18.17 18.83 18.65 18.01 16.69 15.26
PP 5. P 24.78 20.34 16.72 16.78 19.03 19.50 19.12 17.57 16.15 14.93 14.53
PP 6. P 21.83 19.05 17.38 18.85 20.53 19.75 17.75 14.74 12.90 12.99 15.08
PP 7. P 19.29 18.48 18.10 19.58 19.92 17.82 14.65 11.31 11.06 14.25 18.78
PP 8. P 17.77 18.28 18.27 18.96 17.88 14.89 11.58 9.99 12.77 18.00 23.36
PP 9. P 17.75 18.70 18.30 17.57 14.86 11.49 9.77 11.98 17.01 22.92 28.24
PP 10. P 18.95 19.16 17.72 15.25 11.19 8.90 10.92 15.99 21.69 27.35 32.04
PP 11. P 17.96 17.84 16.43 13.95 11.05 12.32 16.60 22.04 27.50 32.70 36.77
PP 12. P 18.06 16.11 13.65 10.92 10.55 15.20 20.85 26.26 31.14 35.42 38.46First, your reported 3DNA groove widths are correct. For other viewers of the Forum, one can easily analyze
1bna with 3DNA as follows:
find_pair 1bna.pdb 1bna.inp
analyze 1bna.inp
# or: find_pair 1bna.pdb stdout | analyze stdin
The main output file
1bna.out (attached) contains 3DNA parameters for the PDB entry
1bna.
Second, the section of 3DNA groove widths also includes a header, providing reference to the 1998 El Hassan and Calladine JMB paper. 3DNA implements the algorithm detailed in "Appendix: Calculation of Major- and Minor-groove Widths" of the paper.
****************************************************************************
Minor and major groove widths: direct P-P distances and refined P-P distances
which take into account the directions of the sugar-phosphate backbones
(Subtract 5.8 Angstrom from the values to take account of the vdw radii
of the phosphate groups, and for comparison with FreeHelix and Curves.)
Ref: M. A. El Hassan and C. R. Calladine (1998). ``Two Distinct Modes of
Protein-induced Bending in DNA.'' J. Mol. Biol., v282, pp331-343.
Minor Groove Major Groove
P-P Refined P-P Refined
1 CG/CG --- --- --- ---
2 GC/GC --- --- --- ---
3 CG/CG 13.8 --- 17.3 ---
4 GA/TC 12.0 12.0 17.3 17.3
5 AA/TT 10.5 10.5 17.5 17.3
6 AT/AT 9.9 9.9 16.8 16.3
7 TT/AA 9.3 9.3 17.4 17.4
8 TC/GA 10.0 9.9 18.5 18.4
9 CG/CG 11.0 --- 17.8 ---
10 GC/GC --- --- --- ---
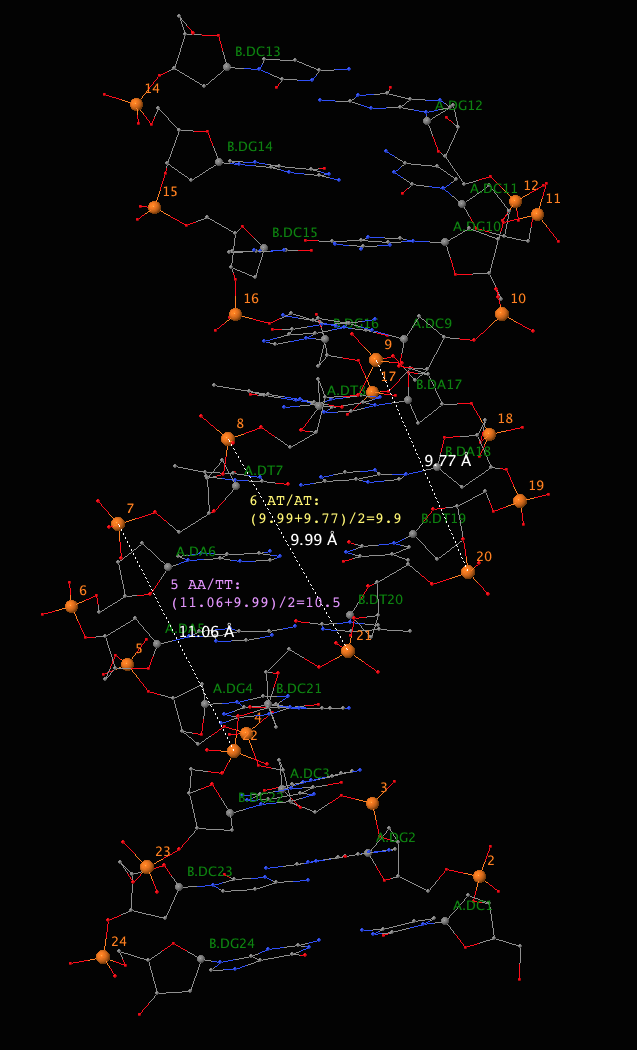
11 CG/CG --- --- --- ---Third, as shown in Eq. (A1) and Figure A1 of the El Hassan and Calladine JMB paper, "Accordingly, we define the minor-groove width for a step such as 4 in Figure A1(a) as the mean of the two marked distances". To make our discussion more concrete, the following image details how the the minor widths for dinucleotide steps 5 (AA/TT) and 6 (AT/AT) as reported in 3DNA come about.
Forth, 3DNA also creates a file named
auxiliary.par (attached) which contains the following section:
Phosphorus-phosphorus distance in Angstroms
1 2 3 4 5 6 7 8 9 10 11 12
G C G C T T A A G C G C
1 C ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ---- ----
2 G 18.0 18.5 18.8 18.3 17.3 17.4 19.6 22.5 27.6 33.0 37.1 ----
3 C 16.8 18.1 19.1 19.0 18.2 17.3 17.6 18.6 22.9 28.5 33.2 ----
4 G 15.3 16.7 18.0 18.6 18.8 18.2 17.5 16.4 18.9 24.0 29.0 ----
5 A 14.5 14.9 16.1 17.6 19.1 19.5 19.0 16.8 16.7 20.3 24.8 ----
6 A 15.1 13.0 12.9 14.7 17.8 19.7 20.5 18.8 17.4 19.0 21.8 ----
7 T 18.8 14.3 11.1 11.3 14.6 17.8 19.9 19.6 18.1 18.5 19.3 ----
8 T 23.4 18.0 12.8 10.0 11.6 14.9 17.9 19.0 18.3 18.3 17.8 ----
9 C 28.2 22.9 17.0 12.0 9.8 11.5 14.9 17.6 18.3 18.7 17.7 ----
10 G 32.0 27.3 21.7 16.0 10.9 8.9 11.2 15.3 17.7 19.2 19.0 ----
11 C 36.8 32.7 27.5 22.0 16.6 12.3 11.0 14.0 16.4 17.8 18.0 ----
12 G 38.5 35.4 31.1 26.3 20.8 15.2 10.6 10.9 13.7 16.1 18.1 ----
As you can see, this output agrees with NUPARM, except for differences in ordering and decimal points.
HTH,
Xiang-Jun


