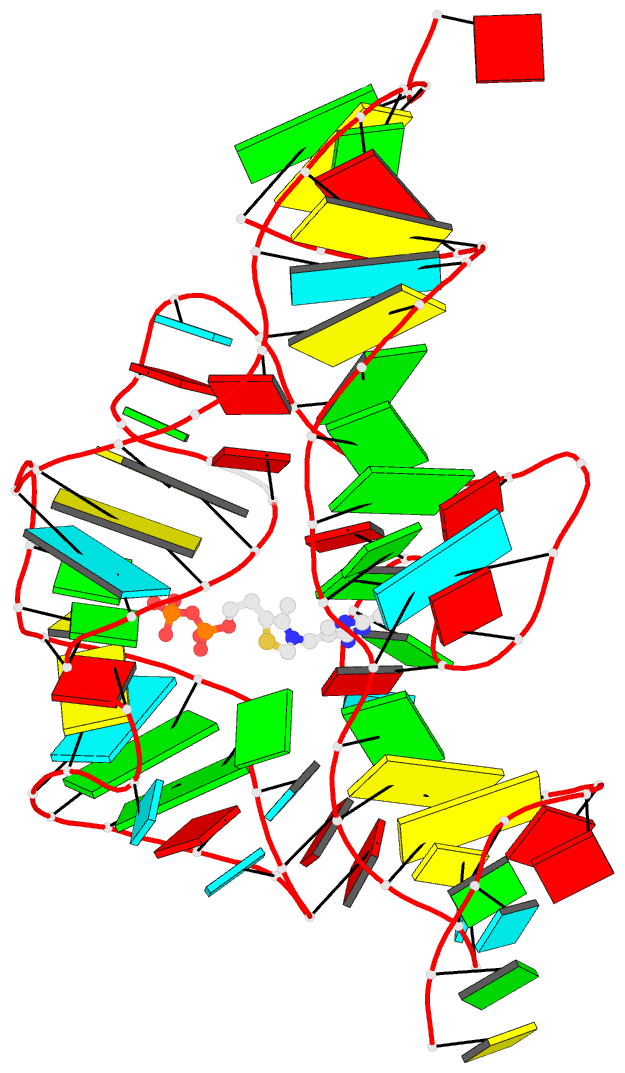
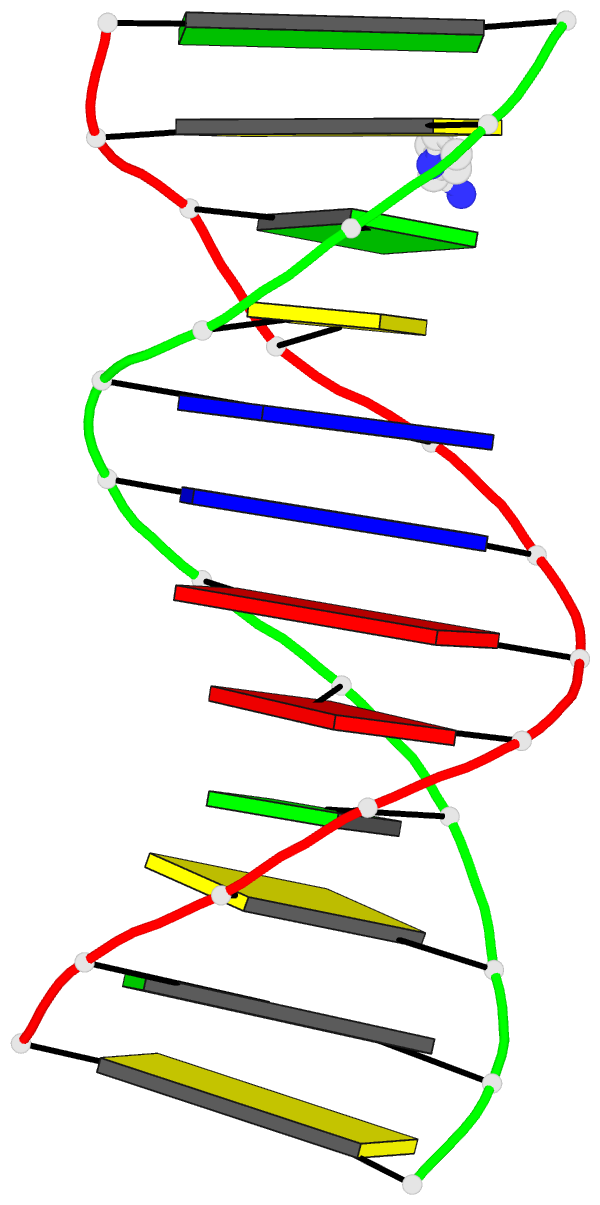
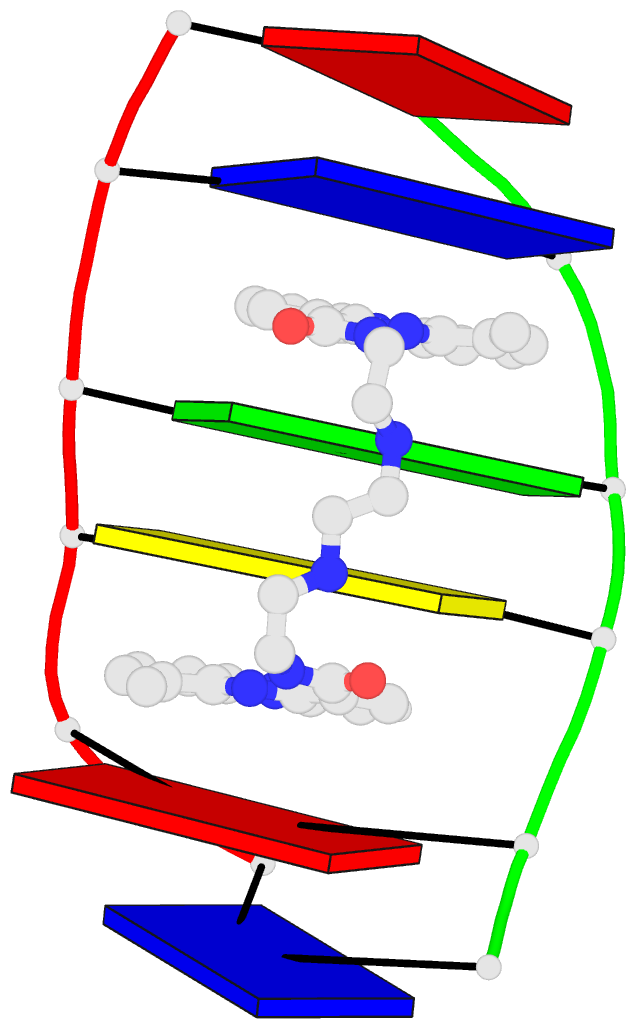
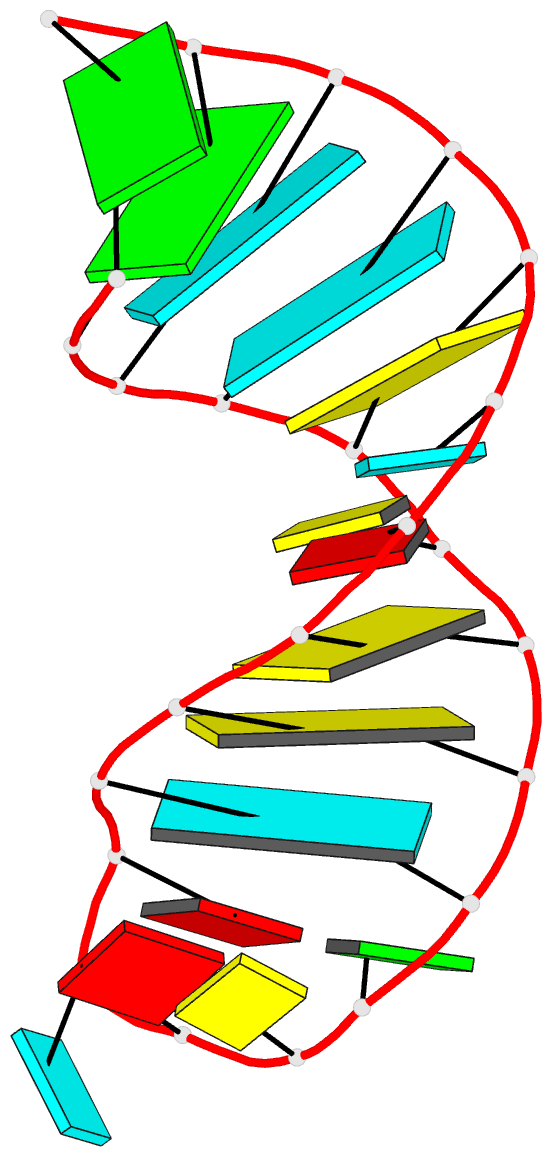
This website
http://skmatic.x3dna.org/ (see screenshot below) aims to showcase DSSR-enabled cartoon-block schematics of nucleic acid structures using PyMOL. It presents a simple interface to browse pre-calculated PDB entries with a set of default settings: long rectangular blocks for Watson-Crick base-pairs, square blocks for G-tetrads in G-quadruplexes, with minor-groove edges in black. Users can also specify an URL to a PDB- or mmCIF-formatted file or upload such an atomic coordinates file directly, and set several common options to customerize to the rendered image.
Moreover, a
web API to DSSR-PyMOL schematics is available to allow for its easy integration into third-party tools.
Input a PDB id
Pre-calculated cartoon-block images together with summary information are available for PDB entries of nucleic-acid-containing structures. Note that gigantic structures like ribosomes that are only represented in mmCIF format are excluded from the resource. The base block images are most effective for small to medium-sized structures.
Here are a few examples:
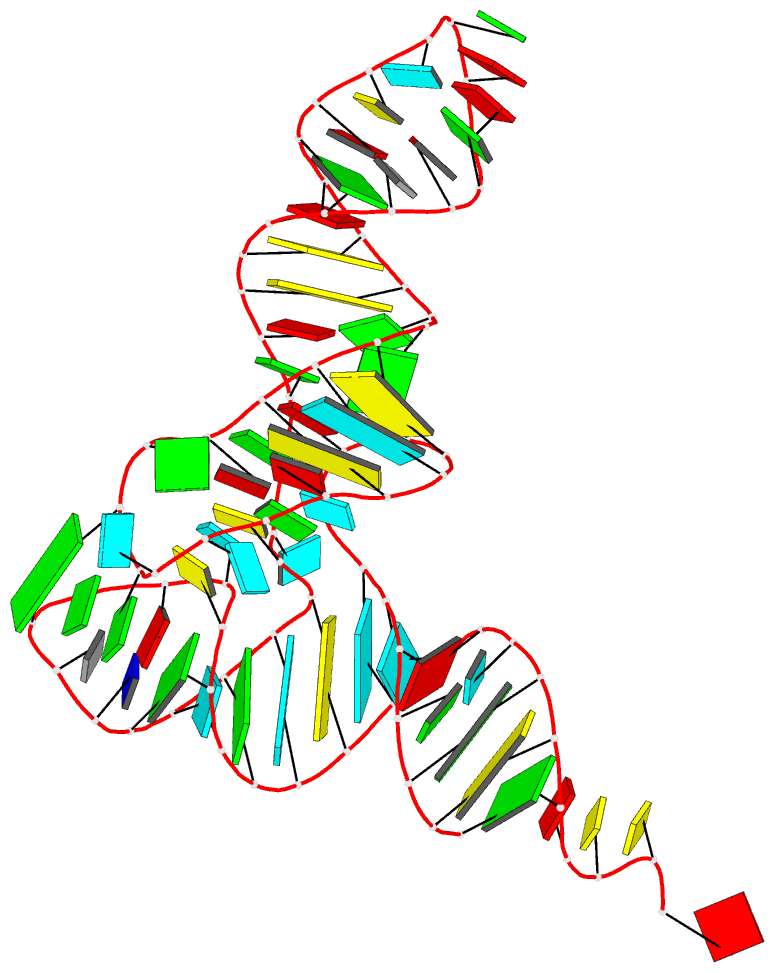
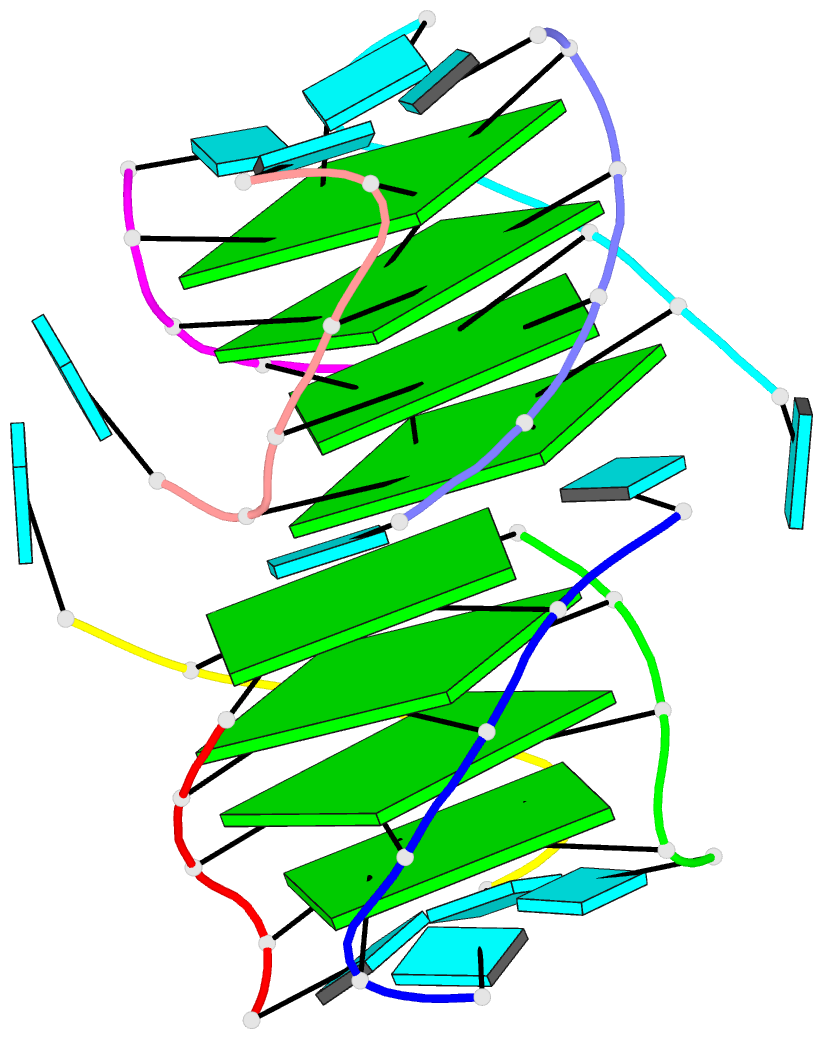
- 1ehz, the crystal structure of yeast phenylalanine trna at 1.93-A resolution
- 2lx1, the major conformation of the internal loop 5'GAGU/3'UGAG
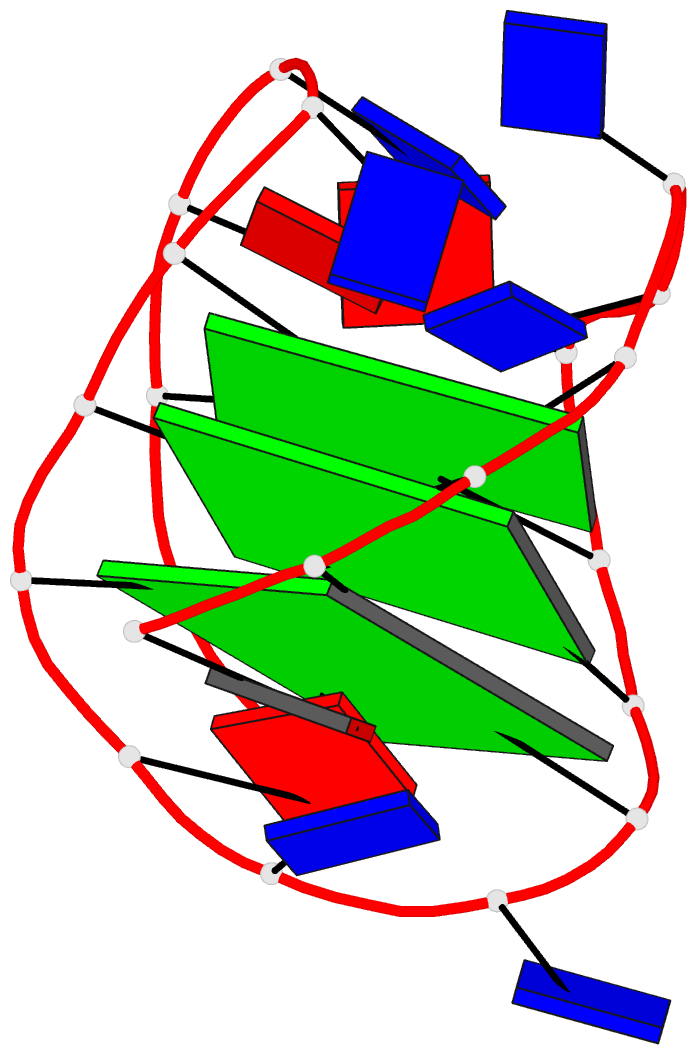
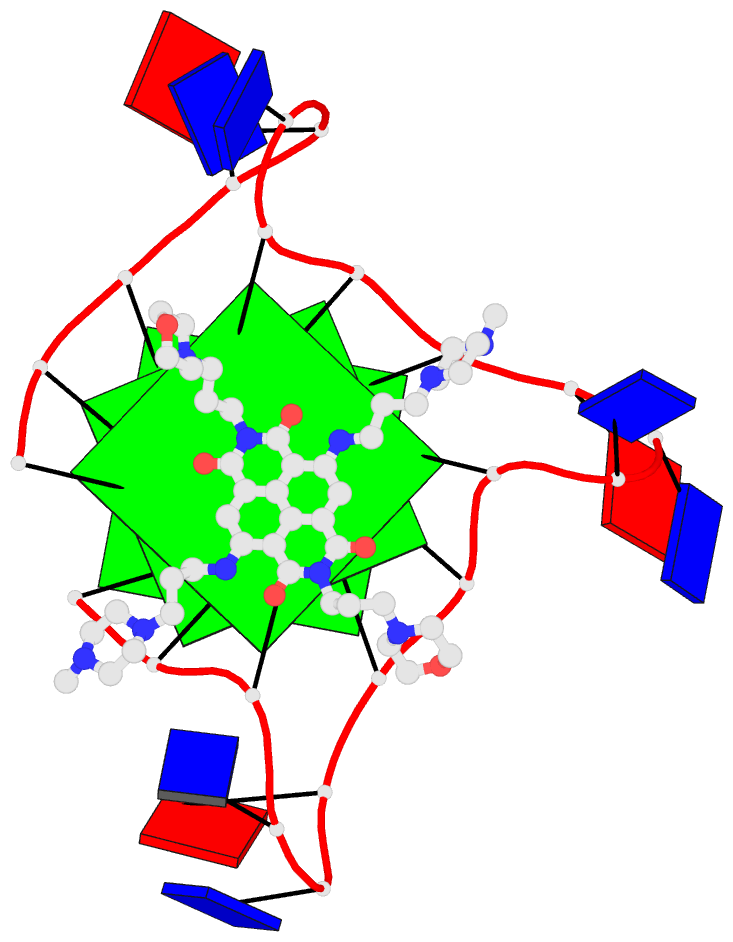
- 2grb, the crystal structure of an RNA quadruplex containing inosine-tetrad
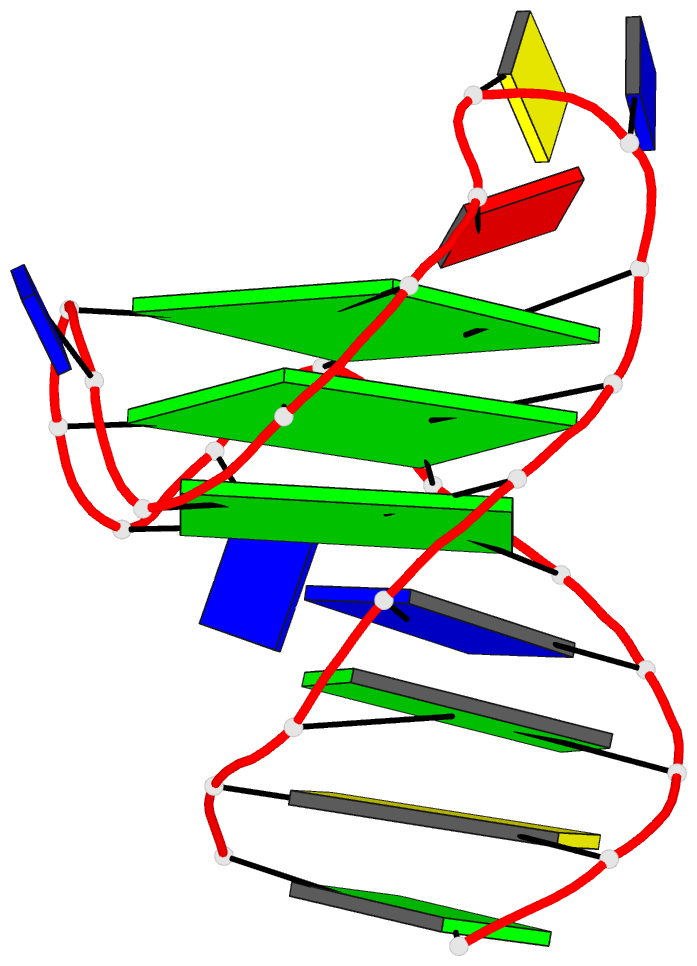
- 4da3, the crystal structure of an intramolecular human telomeric DNA G-quadruplex 21-mer bound by the naphthalene diimide compound MM41
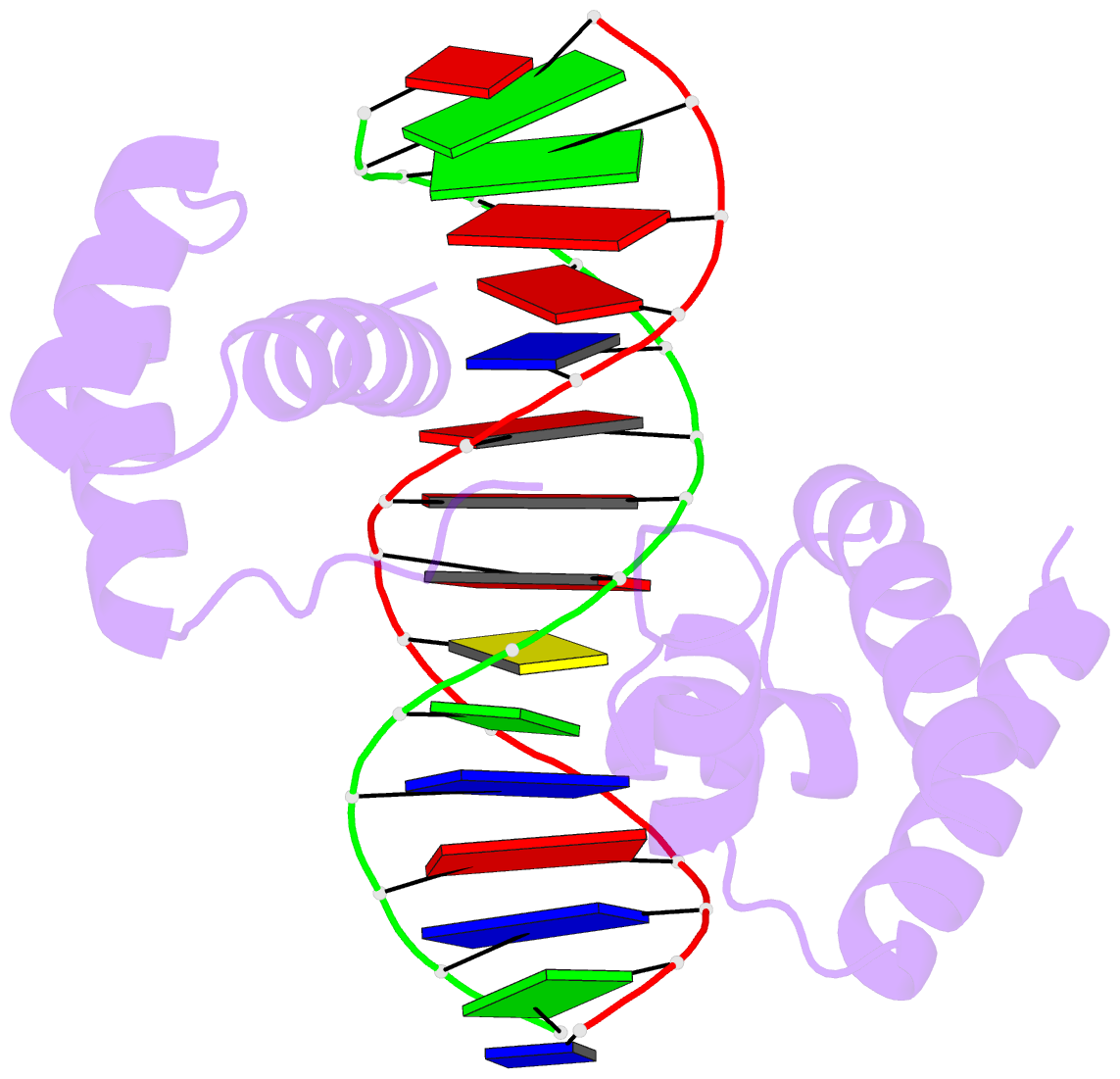
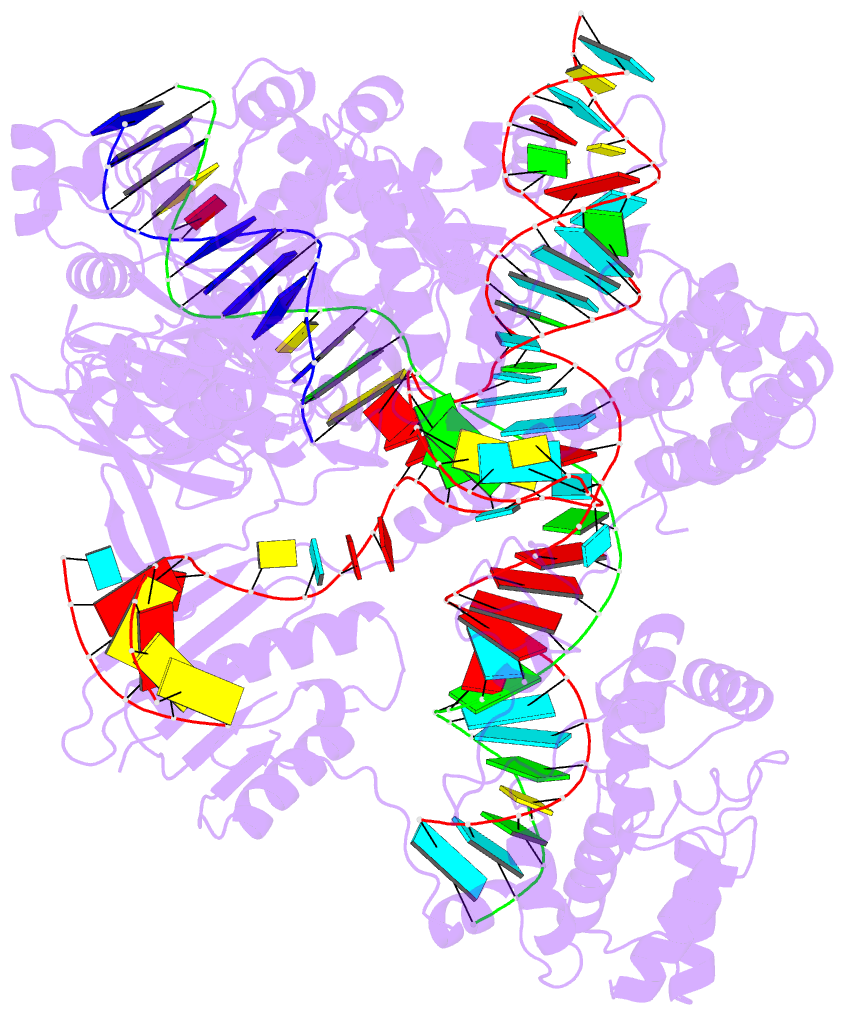
- 1oct, crystal structure of the Oct-1 POU domain bound to an octamer site
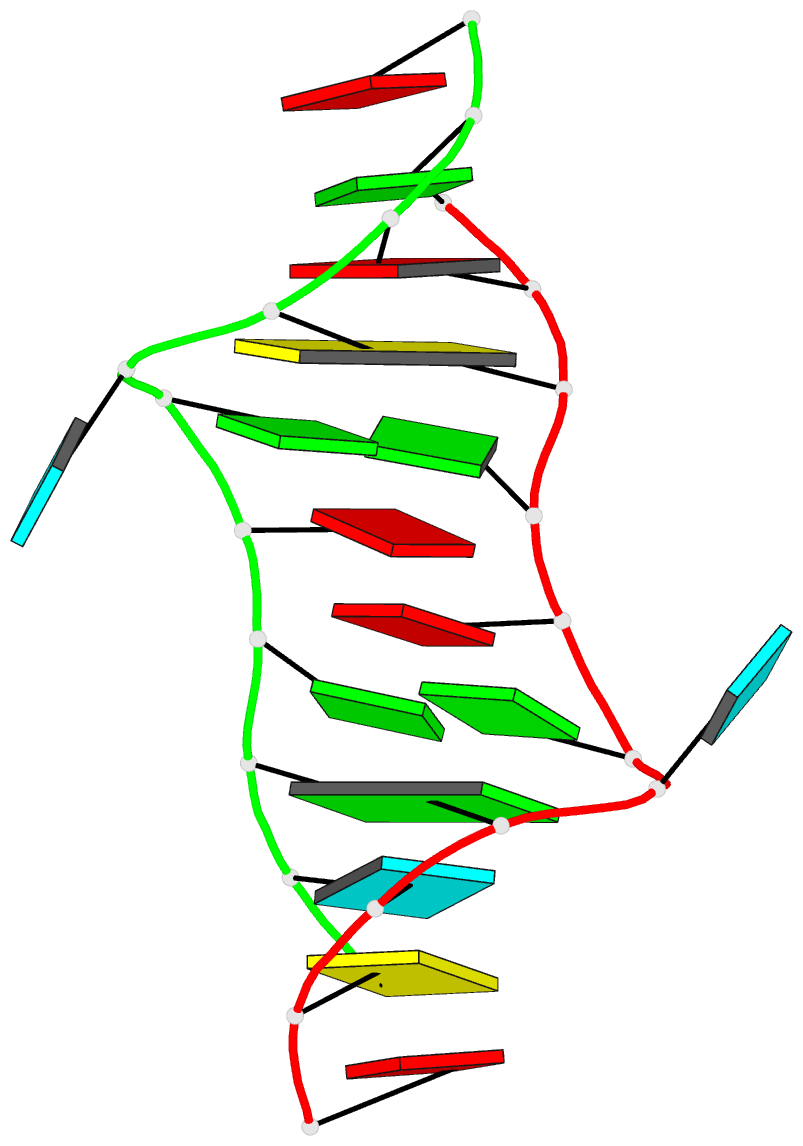
- 2hoj, the crystal structure of an E. coli thi-box riboswitch bound to thiamine pyrophosphate, manganese ions
Each entry is shown with images in six orthogonal perspectives: front, back, right, left, top, bottom. The 'front' image (upper-left in the panel) is oriented into the most-extended view with the DSSR
--blocview option. The corresponding PyMOL session file and PDB coordinate file are available for download. One can also visualize the structure interactively via
3Dmol.js.
Sample PDB entries
Users can browse
random samples of pre-calculated PDB entries. The number should be between 3 and 99, with a default of 12 entries (see below for an example). Simply click the 'Submit' button or the link "
Random samples (3 to 99)" to see random results of 12 entries each time.
Specify an coordinate file
The atomic coordinate file must be in PDB or mmCIF format, and can be optionally gzipped (
.gz). One can either specify an URL to or select a coordinate file. Several common options are available to allow for user customizations.
Web API help message
Usage with 'http' (HTTPie):
http -f http://skmatic.x3dna.org/api [options] url=|model@
http http://skmatic.x3dna.org/api/pdb/pdb_id -- for a pre-calculated PDB entry
http http://skmatic.x3dna.org/api/help -- display this help message
Options:
block_file=styles-in-free-text-format [e.g., block_file=wc-minor]
block_color=nt-selection-and-color [e.g., block_color='A:pink']
block_depth=thickness-of-base-block [e.g., block_depth=1.2]
r3d_file=true-or-FALSE(default) [e.g., r3d_file=true]
raw_xyz=true-or-FALSE(default) [e.g., raw_xyz=true]
Required parameter
url=URL-to-coordinate-file [e.g., url=https://files.rcsb.org/download/1ehz.pdb.gz]
model@coordinate-file [e.g., model@1ehz.cif]
# Only one must be specified. 'url' precedes 'model' when both are specified.
# The coordinate file must be in PDB or PDBx/mmCIF format, optionally gzipped.
Examples
http -f http://skmatic.x3dna.org/api block_file='wc-minor' model@1ehz.cif r3d_file=t
http -f http://skmatic.x3dna.org/api url=https://files.rcsb.org/download/1ehz.pdb.gz -d -o 1ehz.png
http http://skmatic.x3dna.org/api/pdb/1ehz -d -o 1ehz.png
# with 'curl'
curl http://skmatic.x3dna.org/api -F 'model=@1msy.pdb' -F 'block_file=wc-minor' -F 'r3d_file=1'
curl http://skmatic.x3dna.org/api -F 'url=https://files.rcsb.org/download/1ehz.pdb.gz' -o 1ehz.png
curl http://skmatic.x3dna.org/api/pdb/1ehz -o 1ehz.png
Sample images