As of v2.3-2016nov16, the 3DNA
fiber program has added the
--pauling option. This new feature is intended for easy generation of DNA/RNA triplex models based on the historical paper of Pauling and Corey in 1953, titled "
A proposed structure for the nucleic acids". Combined with the existing
--sequence option, 3DNA users can now construct a Pauling triplex model with arbitrary base sequence.
The basic usage is very straightforward, as illustrated in the following list of examples. There are also other variants as well, which may be useful for advanced users.
fiber -pauling triplex-C10C10C10.pdb # default: 10 Cs per strand
fiber -pauling -seq=AAA triplex-A3A3A3.pdb # 3 As per strand
fiber -pauling -seq=AAAA:CCCC:GGGG Pauling-triplex-A4C4G4.pdb
fiber -pauling -seq=ACGGUU,UUGGAC,GGAACC Pauling-triplex-mixed.pdb
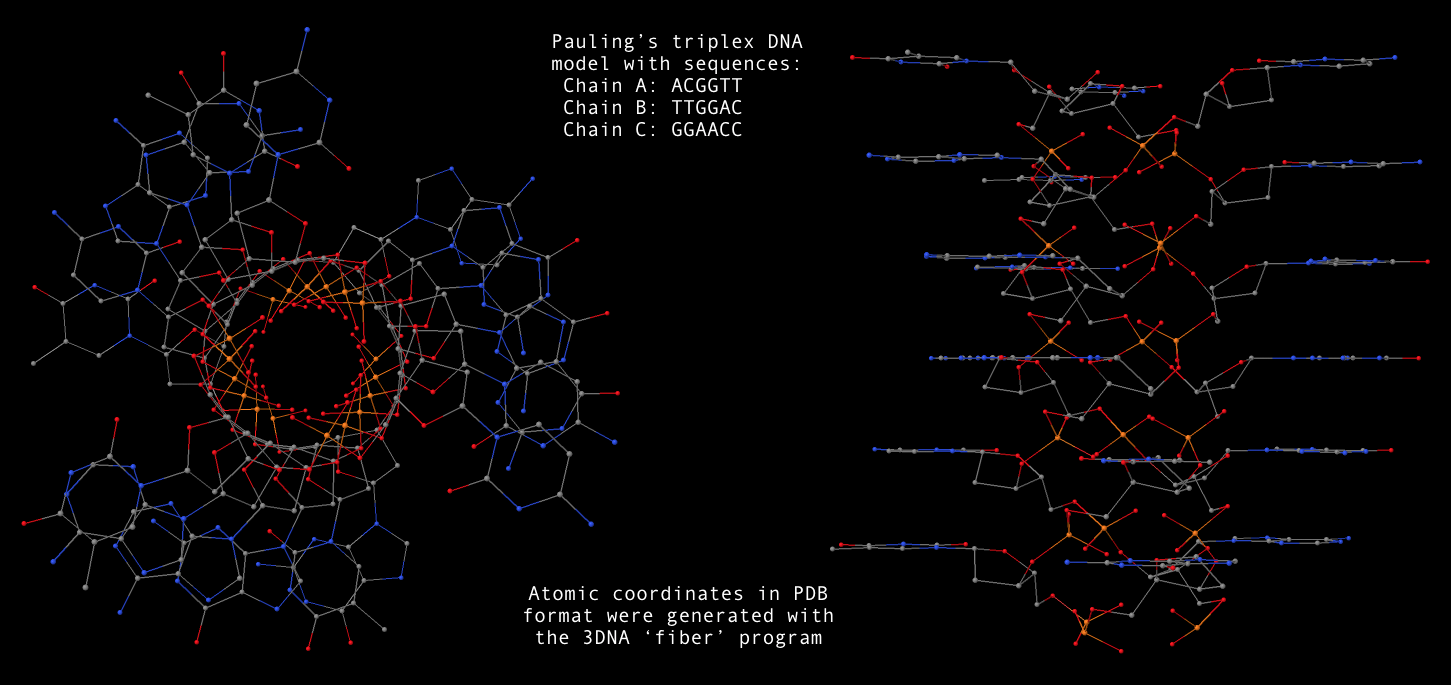
fiber --pauling-dna -seq=ACGGTT,TTGGAC,GGAACC Pauling-triplex-DNA.pdbA sample 3D image for the mixed sequence (last one in the above list,
Pauling-triplex-DNA.pdb) is shown below:
See my blogpost titled "
Pauling's triplex model of nucleic acids is available in 3DNA" for further information.


