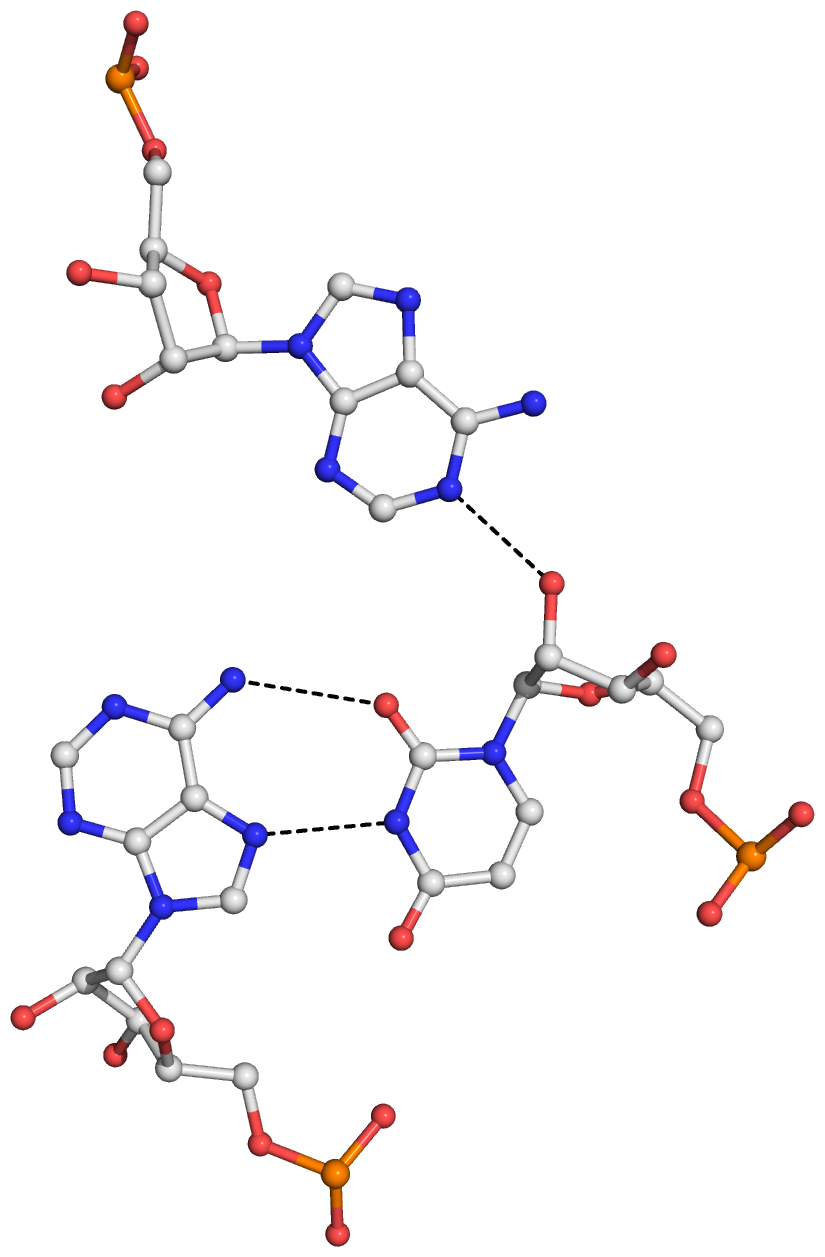
Figure S1: The four base triplets and associated hydrogen bonds (dashed lines) detected by DSSR in yeast tRNAPhe (PDB id: 1ehz). (A) UAA (U8,A14,A21), with a reverse Hoogsteen U8–A14 pair. (B) AUA (A9,U12,A23). (C) gCG (2MG10,C25,G45). (D) CGg (C13,G22,7MG46). Here, the bases in each triplet are listed in sequential order, with the one-letter shorthand form followed by more detailed identifiers in parentheses. In (C) and (D), the lower case ‘g’ represents the shortened name for modified guanine nucleotides 2MG10 and 7MG46, respectively. Note that the triplets and hydrogen bonds match those originally reported by Quigley and Rich for yeast tRNAPhe.
Starting from "
1ehz.pdb" downloaded from RCSB PDB, here is the complete script to get each of the four base-triplet images in png format.
x3dna-dssr -i=1ehz.pdb -o=1ehz.out --prefix=1ehz
ex_str -1 1ehz-multiplets.pdb 1ehz-m1.pdb
x3dna-dssr -i=1ehz-m1.pdb -o=1ehz-m1.pml --hbfile-pymol
pymol -qkc 1ehz-m1.pml
convert -trim +repage -border 10 -bordercolor white 1ehz-m1-pymol.png 1ehz-m1.png
ex_str -2 1ehz-multiplets.pdb 1ehz-m2.pdb
x3dna-dssr -i=1ehz-m2.pdb -o=1ehz-m2.pml --hbfile-pymol
pymol -qkc 1ehz-m2.pml
convert -trim +repage -border 10 -bordercolor white 1ehz-m2-pymol.png 1ehz-m2.png
ex_str -3 1ehz-multiplets.pdb 1ehz-m3.pdb
x3dna-dssr -i=1ehz-m3.pdb -o=1ehz-m3.pml --hbfile-pymol
pymol -qkc 1ehz-m3.pml
convert -trim +repage -border 10 -bordercolor white 1ehz-m3-pymol.png 1ehz-m3.png
ex_str -4 1ehz-multiplets.pdb 1ehz-m4.pdb
x3dna-dssr -i=1ehz-m4.pdb -o=1ehz-m4.pml --hbfile-pymol
pymol -qkc 1ehz-m4.pml
convert -trim +repage -border 10 -bordercolor white 1ehz-m4-pymol.png 1ehz-m4.png
Note:
- The --prefix option makes the auxiliary files having a specified prefix instead of the default "dssr". For example, "dssr-multiplets.pdb" becomes "1ehz-multiplets.pdb".
- The ex_str utility program is from the 3DNA distribution. It is used to extract a specific model from a MODEL/ENDMDL ensemble.
- The DSSR --hbfile-pymol option is used to generate a .pml file with all required settings for rendering in PyMOL.
- The convert program is from ImageMagick that is used here to trim extra white boundaries.
- The multiplet-png images (here four triplets) were combined using InkScape, and annotated, to get the final illustration.
- For completeness, here is the tarball file containing all the data files and the script ("tasks"): supp-fig1-1ehz-multiplets.tar.gz
Here is a sample image generated with the above script: