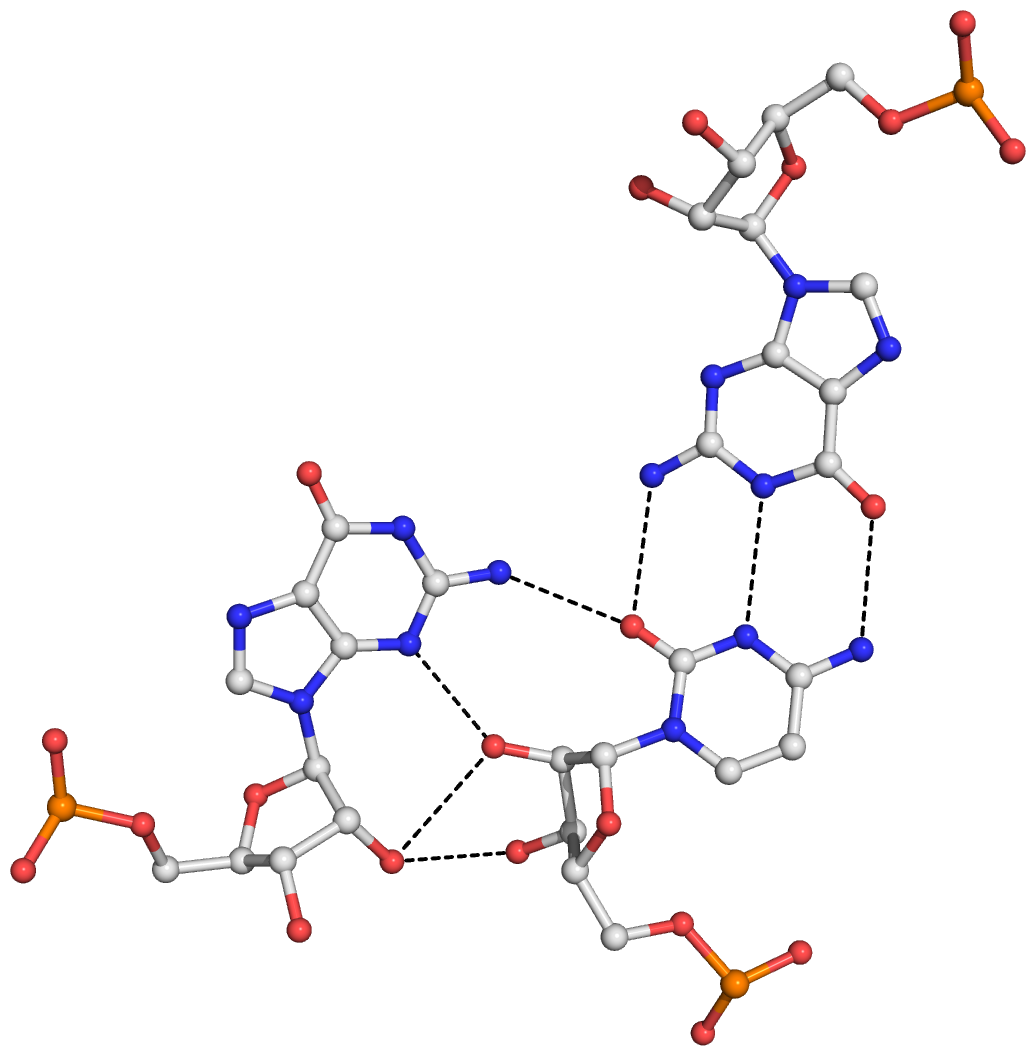
Figure S7: The eight base triplets and associated hydrogen bonds (dashed lines) detected by DSSR in the SAM-I riboswitch (PDB id: 2gis). (A) GCG (G11,C44,G58), with G11 in a similar position and orientation as in a type II A-minor motif. (B) AGC (A12,G43,C59), a type I A-minor motif. (C) AGG (A20,G32,G35). (D) GCA (G22,C30,A61). (E) GCA (G23,C29,A62). (F) AUA (A24,U64,A85), with the isolated, linchpin-like U64-A85 pair. (G) AUa (A45,U57,SAM301), with the SAM adenosine moiety taken as a modified base in forming the triplet. (H) ACG (A46,C47,G56). Note that in (D) and (E), A61 and A62 employ their Watson-Crick edges, rather than the minor-groove edges as in A-minor motifs, to interact with the minor-groove edges of the two consecutive G–C pairs.
Starting from "
2gis.pdb" downloaded from RCSB PDB, here is the complete script to get each of the eight base-triplet images in png format.
x3dna-dssr -i=2gis.pdb -o=2gis.out --prefix=2gis
ex_str -1 2gis-multiplets.pdb 2gis-m1.pdb
x3dna-dssr -i=2gis-m1.pdb -o=2gis-m1.pml --hbfile-pymol
pymol -qkc 2gis-m1.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m1-pymol.png 2gis-m1.png
ex_str -2 2gis-multiplets.pdb 2gis-m2.pdb
x3dna-dssr -i=2gis-m2.pdb -o=2gis-m2.pml --hbfile-pymol
pymol -qkc 2gis-m2.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m2-pymol.png 2gis-m2.png
ex_str -3 2gis-multiplets.pdb 2gis-m3.pdb
x3dna-dssr -i=2gis-m3.pdb -o=2gis-m3.pml --hbfile-pymol
pymol -qkc 2gis-m3.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m3-pymol.png 2gis-m3.png
ex_str -4 2gis-multiplets.pdb 2gis-m4.pdb
x3dna-dssr -i=2gis-m4.pdb -o=2gis-m4.pml --hbfile-pymol
pymol -qkc 2gis-m4.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m4-pymol.png 2gis-m4.png
ex_str -5 2gis-multiplets.pdb 2gis-m5.pdb
x3dna-dssr -i=2gis-m5.pdb -o=2gis-m5.pml --hbfile-pymol
pymol -qkc 2gis-m5.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m5-pymol.png 2gis-m5.png
ex_str -6 2gis-multiplets.pdb 2gis-m6.pdb
x3dna-dssr -i=2gis-m6.pdb -o=2gis-m6.pml --hbfile-pymol
pymol -qkc 2gis-m6.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m6-pymol.png 2gis-m6.png
ex_str -7 2gis-multiplets.pdb 2gis-m7.pdb
x3dna-dssr -i=2gis-m7.pdb -o=2gis-m7.pml --hbfile-pymol
pymol -qkc 2gis-m7.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m7-pymol.png 2gis-m7.png
ex_str -8 2gis-multiplets.pdb 2gis-m8.pdb
x3dna-dssr -i=2gis-m8.pdb -o=2gis-m8.pml --hbfile-pymol
pymol -qkc 2gis-m8.pml
convert -trim +repage -border 10 -bordercolor white 2gis-m8-pymol.png 2gis-m8.png
Note:
- The --prefix option makes the auxiliary files having a specified prefix instead of the default "dssr". For example, "dssr-multiplets.pdb" becomes "2gis-multiplets.pdb".
- The ex_str utility program is from the 3DNA distribution. It is used to extract a specific model from a MODEL/ENDMDL ensemble.
- The DSSR --hbfile-pymol option is used to generate a .pml file with all required settings for rendering in PyMOL.
- The convert program is from ImageMagick that is used here to trim extra white boundaries.
- The multiplet-png images (here four triplets) were combined using InkScape, and annotated, to get the final illustration.
- For completeness, here is the tarball file containing all the data files and the script ("tasks"): supp-fig7-2gis-multiplets.tar.gz
Here is a sample image generated with the above script: