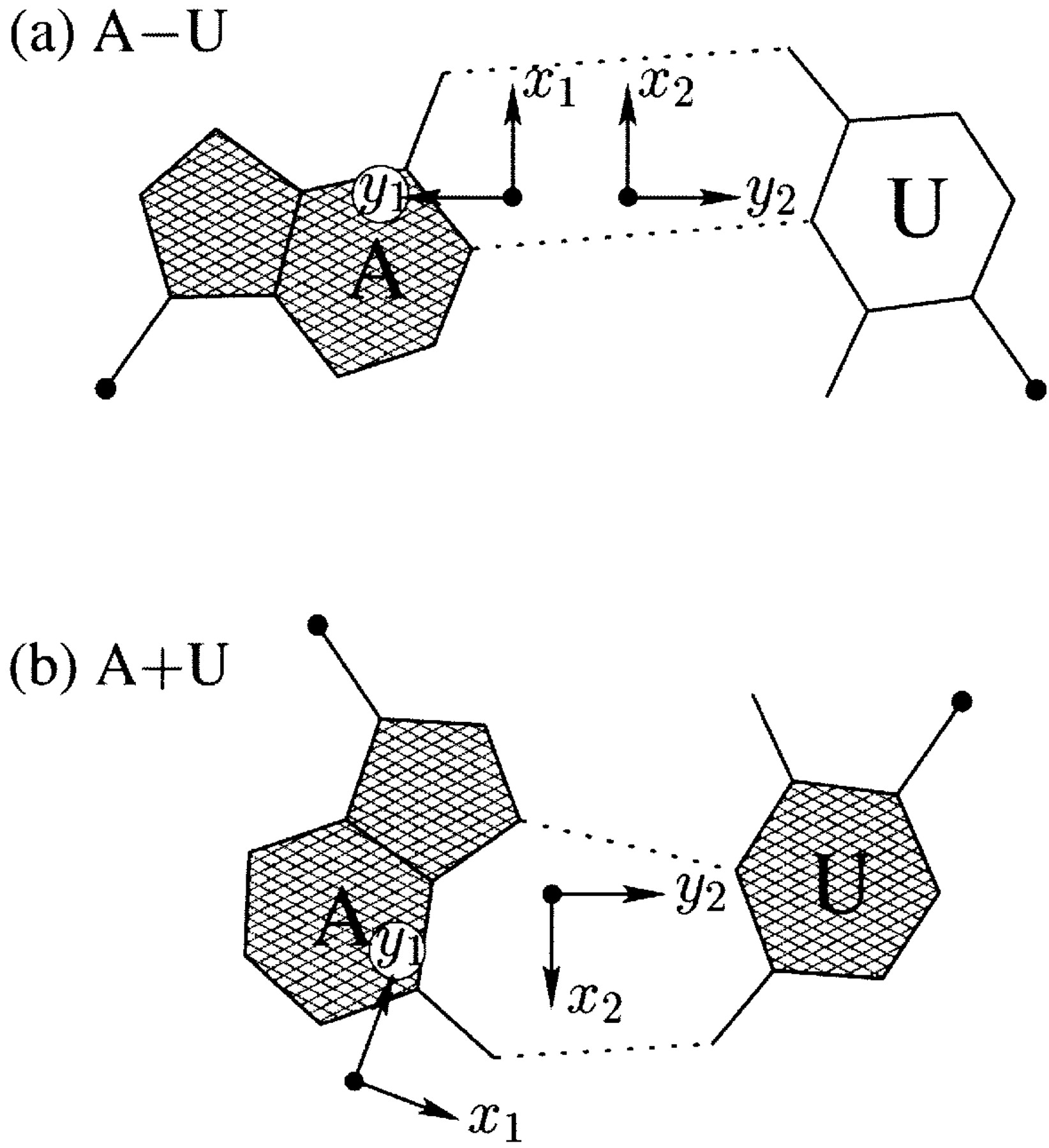
Thanks for using w3DNA, a web-service hosted and supported by the Olson laboratory at Rutgers University. w3DNA aims to make commonly/routinely used 3DNA functionality easily available. To taking full advantage of 3DNA, the standard command-line version is the way to go. Also, w3DNA is still using 3DNA v2.0, while the the command-line version is v2.1.
The issue you experienced for PDB entries
2o8b and
2o8c, i.e., some step parameters are designated as "
----", is due to the kink in the two structures. Thus,
find_pair now takes each DNA as two helical regions instead of a continuous helix, see below:
find_pair 2o8b.pdb 2o8b.inp
# contents of 2o8b.inp
2o8b.pdb
2o8b.out
2 # duplex
15 # number of base-pairs
1 1 # explicit bp numbering/hetero atoms
1 30 0 # 1 | ....>E:...1_:[.DG]G-----C[.DC]:..30_:F<.... 1.14 0.36 14.09 9.30 -0.44
2 29 0 # 2 | ....>E:...2_:[.DA]A-----T[.DT]:..29_:F<.... 0.89 0.82 26.82 9.09 -1.13
3 28 0 # 3 | ....>E:...3_:[.DA]A-----T[.DT]:..28_:F<.... 0.23 0.03 18.38 9.24 -3.78
4 27 0 # 4 | ....>E:...4_:[.DC]C-----G[.DG]:..27_:F<.... 0.78 0.19 8.59 8.93 -3.40
5 26 0 # 5 | ....>E:...5_:[.DC]C-----G[.DG]:..26_:F<.... 0.56 0.29 17.33 9.24 -3.00
6 25 0 # 6 | ....>E:...6_:[.DG]G-----C[.DC]:..25_:F<.... 0.29 0.27 19.28 9.01 -3.20
7 24 9 # 7 x ....>E:...7_:[.DC]C-----G[.DG]:..24_:F<.... 0.22 0.01 24.18 9.04 -3.55
8 23 0 # 8 | ....>E:...8_:[.DG]G-**--T[.DT]:..23_:F<.... 5.22 0.31 43.28 9.87 7.00
9 22 0 # 9 | ....>E:...9_:[.DC]C-----G[.DG]:..22_:F<.... 0.44 0.33 20.98 8.84 -2.85
10 21 0 # 10 | ....>E:..10_:[.DG]G-----C[.DC]:..21_:F<.... 0.41 0.39 10.80 9.05 -3.28
11 20 0 # 11 | ....>E:..11_:[.DC]C-----G[.DG]:..20_:F<.... 0.26 0.01 12.86 9.06 -4.07
12 19 0 # 12 | ....>E:..12_:[.DT]T-----A[.DA]:..19_:F<.... 0.85 0.47 11.88 8.95 -2.62
13 18 0 # 13 | ....>E:..13_:[.DA]A-----T[.DT]:..18_:F<.... 0.53 0.18 9.30 8.85 -3.65
14 17 0 # 14 | ....>E:..14_:[.DG]G-----C[.DC]:..17_:F<.... 1.17 0.94 21.28 8.97 -0.89
15 16 0 # 15 | ....>E:..15_:[.DG]G-----C[.DC]:..16_:F<.... 1.17 0.05 37.85 8.66 -1.83
##### Base-pair criteria used: 4.00 0.00 15.00 2.50 65.00 4.50 7.50 [ O N]
##### 1 non-Watson-Crick base-pair, and 2 helices (0 isolated bps)
##### Helix #1 (7 ): 1 - 7
##### Helix #2 (8 ): 8 - 15
You can fix the problem by either changing 9 for bp #7 to 0, or running
analyze with the
-c option:
analyze -c 2o8b.inpIn 3DNA, lower case a/c/g/t/u is used for modified bases of A/C/G/T/U respectively. For example, in
2o8c,
6OG is assigned as
g.
HTH,
Xiang-Jun